Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for SOX8
Z-value: 1.33
Transcription factors associated with SOX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX8
|
ENSG00000005513.9 | SOX8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX8 | hg19_v2_chr16_+_1031762_1031808 | -0.39 | 3.4e-01 | Click! |
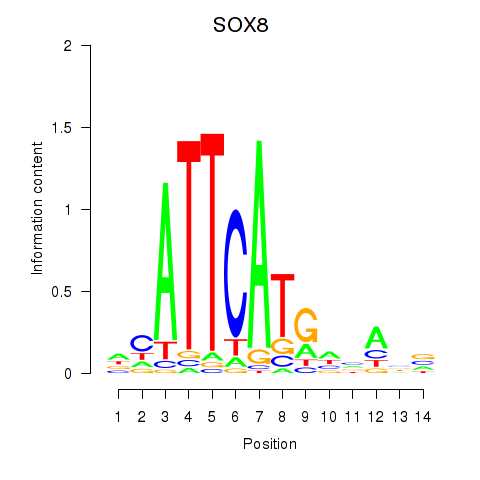
Activity profile of SOX8 motif
Sorted Z-values of SOX8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_61442629 | 2.01 |
ENST00000398019.2 ENST00000540675.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_152956549 | 1.65 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr15_+_43885252 | 1.53 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43985084 | 1.49 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr6_-_136788001 | 1.48 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr1_+_153330322 | 1.37 |
ENST00000368738.3 |
S100A9 |
S100 calcium binding protein A9 |
| chr11_+_12308447 | 1.23 |
ENST00000256186.2 |
MICALCL |
MICAL C-terminal like |
| chr5_+_140227048 | 1.17 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr14_+_75745477 | 1.14 |
ENST00000303562.4 ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr1_-_153029980 | 0.88 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr19_-_49567124 | 0.85 |
ENST00000301411.3 |
NTF4 |
neurotrophin 4 |
| chr2_+_102615416 | 0.83 |
ENST00000393414.2 |
IL1R2 |
interleukin 1 receptor, type II |
| chr5_+_140186647 | 0.83 |
ENST00000512229.2 ENST00000356878.4 ENST00000530339.1 |
PCDHA4 |
protocadherin alpha 4 |
| chr18_-_47376197 | 0.82 |
ENST00000592688.1 |
MYO5B |
myosin VB |
| chr17_-_8021710 | 0.77 |
ENST00000380149.1 ENST00000448843.2 |
ALOXE3 |
arachidonate lipoxygenase 3 |
| chr4_-_47983519 | 0.74 |
ENST00000358519.4 ENST00000544810.1 ENST00000402813.3 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chr11_-_125366089 | 0.72 |
ENST00000366139.3 ENST00000278919.3 |
FEZ1 |
fasciculation and elongation protein zeta 1 (zygin I) |
| chr14_+_96722539 | 0.71 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr11_+_18154059 | 0.68 |
ENST00000531264.1 |
MRGPRX3 |
MAS-related GPR, member X3 |
| chr8_-_91095099 | 0.67 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chr12_+_29376673 | 0.66 |
ENST00000547116.1 |
FAR2 |
fatty acyl CoA reductase 2 |
| chrX_-_117107542 | 0.65 |
ENST00000371878.1 |
KLHL13 |
kelch-like family member 13 |
| chr22_-_20255212 | 0.65 |
ENST00000416372.1 |
RTN4R |
reticulon 4 receptor |
| chr13_+_97928395 | 0.64 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr18_-_78005231 | 0.63 |
ENST00000470488.2 ENST00000353265.3 |
PARD6G |
par-6 family cell polarity regulator gamma |
| chr4_+_89299994 | 0.60 |
ENST00000264346.7 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr12_+_20848282 | 0.56 |
ENST00000545604.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr16_+_618837 | 0.54 |
ENST00000409439.2 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr12_+_20848377 | 0.53 |
ENST00000540354.1 ENST00000266509.2 ENST00000381552.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chrX_-_117107680 | 0.53 |
ENST00000447671.2 ENST00000262820.3 |
KLHL13 |
kelch-like family member 13 |
| chr11_+_59522837 | 0.51 |
ENST00000437946.2 |
STX3 |
syntaxin 3 |
| chr12_+_29376592 | 0.50 |
ENST00000182377.4 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr14_+_96722152 | 0.49 |
ENST00000216629.6 |
BDKRB1 |
bradykinin receptor B1 |
| chr11_+_128563652 | 0.49 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_-_78809950 | 0.48 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr1_+_70876926 | 0.48 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr4_-_77328458 | 0.47 |
ENST00000388914.3 ENST00000434846.2 |
CCDC158 |
coiled-coil domain containing 158 |
| chr5_+_89770664 | 0.47 |
ENST00000503973.1 ENST00000399107.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr5_+_89770696 | 0.46 |
ENST00000504930.1 ENST00000514483.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr3_-_190167571 | 0.44 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr3_-_165555200 | 0.44 |
ENST00000479451.1 ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE |
butyrylcholinesterase |
| chr8_+_21915368 | 0.43 |
ENST00000265800.5 ENST00000517418.1 |
DMTN |
dematin actin binding protein |
| chr12_+_113354341 | 0.43 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr11_+_128563948 | 0.42 |
ENST00000534087.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr21_-_47352477 | 0.41 |
ENST00000593412.1 |
PRED62 |
Uncharacterized protein |
| chr18_-_53253323 | 0.41 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chr3_+_142342228 | 0.40 |
ENST00000337777.3 |
PLS1 |
plastin 1 |
| chr12_-_123201337 | 0.40 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chrX_+_41548220 | 0.39 |
ENST00000378142.4 |
GPR34 |
G protein-coupled receptor 34 |
| chr9_+_706842 | 0.38 |
ENST00000382293.3 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr17_+_1646130 | 0.37 |
ENST00000453066.1 ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr3_-_52090461 | 0.37 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr8_-_86253888 | 0.36 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr18_-_53253112 | 0.36 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr19_+_1524072 | 0.36 |
ENST00000454744.2 |
PLK5 |
polo-like kinase 5 |
| chr1_-_116383738 | 0.36 |
ENST00000320238.3 |
NHLH2 |
nescient helix loop helix 2 |
| chr10_-_1779663 | 0.36 |
ENST00000381312.1 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr15_-_65715401 | 0.35 |
ENST00000352385.2 |
IGDCC4 |
immunoglobulin superfamily, DCC subclass, member 4 |
| chr12_+_133066137 | 0.35 |
ENST00000434748.2 |
FBRSL1 |
fibrosin-like 1 |
| chr17_-_38938786 | 0.35 |
ENST00000301656.3 |
KRT27 |
keratin 27 |
| chr4_+_155484155 | 0.35 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr3_-_193272588 | 0.34 |
ENST00000295548.3 |
ATP13A4 |
ATPase type 13A4 |
| chr8_+_107738343 | 0.32 |
ENST00000521592.1 |
OXR1 |
oxidation resistance 1 |
| chr12_-_123187890 | 0.31 |
ENST00000328880.5 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
| chr8_-_20040638 | 0.31 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr1_+_13516066 | 0.31 |
ENST00000332192.6 |
PRAMEF21 |
PRAME family member 21 |
| chr4_+_74347400 | 0.31 |
ENST00000226355.3 |
AFM |
afamin |
| chr14_-_21562648 | 0.30 |
ENST00000555270.1 |
ZNF219 |
zinc finger protein 219 |
| chr1_+_111415757 | 0.29 |
ENST00000429072.2 ENST00000271324.5 |
CD53 |
CD53 molecule |
| chr3_-_141719195 | 0.29 |
ENST00000397991.4 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr8_+_1993173 | 0.29 |
ENST00000523438.1 |
MYOM2 |
myomesin 2 |
| chr19_+_52076425 | 0.29 |
ENST00000436511.2 |
ZNF175 |
zinc finger protein 175 |
| chr7_-_80551671 | 0.29 |
ENST00000419255.2 ENST00000544525.1 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr1_-_109935819 | 0.28 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr4_+_95972822 | 0.28 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr8_+_1993152 | 0.28 |
ENST00000262113.4 |
MYOM2 |
myomesin 2 |
| chr12_+_45686457 | 0.28 |
ENST00000441606.2 |
ANO6 |
anoctamin 6 |
| chr10_-_75676400 | 0.27 |
ENST00000412307.2 |
C10orf55 |
chromosome 10 open reading frame 55 |
| chr12_+_21207503 | 0.27 |
ENST00000545916.1 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr12_+_56325812 | 0.27 |
ENST00000394147.1 ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr19_-_47287990 | 0.27 |
ENST00000593713.1 ENST00000598022.1 ENST00000434726.2 |
SLC1A5 |
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr3_-_196911002 | 0.26 |
ENST00000452595.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr3_+_97887544 | 0.26 |
ENST00000356526.2 |
OR5H15 |
olfactory receptor, family 5, subfamily H, member 15 |
| chr16_+_82068830 | 0.26 |
ENST00000199936.4 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr6_-_27100529 | 0.26 |
ENST00000607124.1 ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ |
histone cluster 1, H2bj |
| chr6_+_37012607 | 0.25 |
ENST00000423336.1 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr1_-_222763240 | 0.25 |
ENST00000352967.4 ENST00000391882.1 ENST00000543857.1 |
TAF1A |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr4_-_69434245 | 0.25 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr2_+_11674213 | 0.25 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr1_+_110577229 | 0.24 |
ENST00000369795.3 ENST00000369794.2 |
STRIP1 |
striatin interacting protein 1 |
| chr10_-_131762105 | 0.24 |
ENST00000368648.3 ENST00000355311.5 |
EBF3 |
early B-cell factor 3 |
| chr19_-_45681482 | 0.24 |
ENST00000592647.1 ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A |
trafficking protein particle complex 6A |
| chr8_+_107738240 | 0.24 |
ENST00000449762.2 ENST00000297447.6 |
OXR1 |
oxidation resistance 1 |
| chr18_-_29264467 | 0.24 |
ENST00000383131.3 ENST00000237019.7 |
B4GALT6 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr4_-_75695366 | 0.24 |
ENST00000512743.1 |
BTC |
betacellulin |
| chr16_+_22308717 | 0.23 |
ENST00000299853.5 ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E |
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr3_-_193272741 | 0.23 |
ENST00000392443.3 |
ATP13A4 |
ATPase type 13A4 |
| chr3_+_137906154 | 0.22 |
ENST00000466749.1 ENST00000358441.2 ENST00000489213.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr18_+_61575200 | 0.22 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chrX_+_41548259 | 0.22 |
ENST00000378138.5 |
GPR34 |
G protein-coupled receptor 34 |
| chr16_-_75498553 | 0.22 |
ENST00000569276.1 ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A RP11-77K12.1 |
transmembrane protein 170A Uncharacterized protein |
| chr19_+_19144384 | 0.22 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr11_+_43702322 | 0.21 |
ENST00000395700.4 |
HSD17B12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr16_-_30122717 | 0.21 |
ENST00000566613.1 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr18_-_29264669 | 0.20 |
ENST00000306851.5 |
B4GALT6 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr6_-_150217195 | 0.19 |
ENST00000532335.1 |
RAET1E |
retinoic acid early transcript 1E |
| chr8_-_82359662 | 0.19 |
ENST00000519260.1 ENST00000256103.2 |
PMP2 |
peripheral myelin protein 2 |
| chr4_-_100356291 | 0.19 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr13_-_47012325 | 0.19 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr16_+_57769635 | 0.18 |
ENST00000379661.3 ENST00000562592.1 ENST00000566726.1 |
KATNB1 |
katanin p80 (WD repeat containing) subunit B 1 |
| chr18_-_3845293 | 0.18 |
ENST00000400145.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr1_+_26496362 | 0.18 |
ENST00000374266.5 ENST00000270812.5 |
ZNF593 |
zinc finger protein 593 |
| chr1_-_247921982 | 0.18 |
ENST00000408896.2 |
OR1C1 |
olfactory receptor, family 1, subfamily C, member 1 |
| chr5_+_156887027 | 0.18 |
ENST00000435489.2 ENST00000311946.7 |
NIPAL4 |
NIPA-like domain containing 4 |
| chr12_-_15104040 | 0.18 |
ENST00000541644.1 ENST00000545895.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr16_+_87985029 | 0.17 |
ENST00000439677.1 ENST00000286122.7 ENST00000355163.5 ENST00000454563.1 ENST00000479780.2 ENST00000393208.2 ENST00000412691.1 ENST00000355022.4 |
BANP |
BTG3 associated nuclear protein |
| chr1_-_231005310 | 0.17 |
ENST00000470540.1 |
C1orf198 |
chromosome 1 open reading frame 198 |
| chr20_+_48807351 | 0.17 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chrX_-_132887729 | 0.17 |
ENST00000406757.2 |
GPC3 |
glypican 3 |
| chr4_+_155484103 | 0.17 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr21_-_32185570 | 0.17 |
ENST00000329621.4 |
KRTAP8-1 |
keratin associated protein 8-1 |
| chr3_-_123339418 | 0.17 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr7_+_139478030 | 0.17 |
ENST00000425687.1 ENST00000263552.6 ENST00000438104.1 ENST00000336425.5 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr11_+_60995849 | 0.17 |
ENST00000537932.1 |
PGA4 |
pepsinogen 4, group I (pepsinogen A) |
| chr12_+_48876275 | 0.17 |
ENST00000314014.2 |
C12orf54 |
chromosome 12 open reading frame 54 |
| chr14_+_39583427 | 0.17 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr10_+_88414298 | 0.16 |
ENST00000372071.2 |
OPN4 |
opsin 4 |
| chrX_+_41306575 | 0.16 |
ENST00000342595.2 ENST00000378220.1 |
NYX |
nyctalopin |
| chr9_-_71629010 | 0.16 |
ENST00000377276.2 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
| chr8_+_52730143 | 0.16 |
ENST00000415643.1 |
AC090186.1 |
Uncharacterized protein |
| chr2_-_14541060 | 0.16 |
ENST00000418420.1 ENST00000417751.1 |
LINC00276 |
long intergenic non-protein coding RNA 276 |
| chr9_-_128246769 | 0.16 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr3_+_35683651 | 0.16 |
ENST00000187397.4 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr2_+_74425689 | 0.16 |
ENST00000394053.2 ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr16_+_29690358 | 0.16 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr16_+_21244986 | 0.15 |
ENST00000311620.5 |
ANKS4B |
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr11_+_5474638 | 0.15 |
ENST00000341449.2 |
OR51I2 |
olfactory receptor, family 51, subfamily I, member 2 |
| chr13_+_98794810 | 0.15 |
ENST00000595437.1 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr6_-_159421198 | 0.15 |
ENST00000252655.1 ENST00000297262.3 ENST00000367069.2 |
RSPH3 |
radial spoke 3 homolog (Chlamydomonas) |
| chr1_-_111148241 | 0.15 |
ENST00000440270.1 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr13_+_43355683 | 0.15 |
ENST00000537894.1 |
FAM216B |
family with sequence similarity 216, member B |
| chr6_-_52668605 | 0.14 |
ENST00000334575.5 |
GSTA1 |
glutathione S-transferase alpha 1 |
| chr9_-_130661916 | 0.14 |
ENST00000373142.1 ENST00000373146.1 ENST00000373144.3 |
ST6GALNAC6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr6_-_117150198 | 0.14 |
ENST00000310357.3 ENST00000368549.3 ENST00000530250.1 |
GPRC6A |
G protein-coupled receptor, family C, group 6, member A |
| chr1_-_9811600 | 0.14 |
ENST00000435891.1 |
CLSTN1 |
calsyntenin 1 |
| chr6_-_32498046 | 0.13 |
ENST00000374975.3 |
HLA-DRB5 |
major histocompatibility complex, class II, DR beta 5 |
| chr2_+_219646462 | 0.13 |
ENST00000258415.4 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr12_-_9760482 | 0.13 |
ENST00000229402.3 |
KLRB1 |
killer cell lectin-like receptor subfamily B, member 1 |
| chr11_+_9482551 | 0.13 |
ENST00000438144.2 ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143 |
zinc finger protein 143 |
| chr3_-_129147432 | 0.13 |
ENST00000503957.1 ENST00000505956.1 ENST00000326085.3 |
EFCAB12 |
EF-hand calcium binding domain 12 |
| chr3_+_154958715 | 0.12 |
ENST00000462531.1 ENST00000490497.1 |
RP11-451G4.1 |
RP11-451G4.1 |
| chr1_+_13742808 | 0.12 |
ENST00000602960.1 |
PRAMEF20 |
PRAME family member 20 |
| chr5_+_175288631 | 0.12 |
ENST00000509837.1 |
CPLX2 |
complexin 2 |
| chr9_+_127624387 | 0.12 |
ENST00000353214.2 |
ARPC5L |
actin related protein 2/3 complex, subunit 5-like |
| chr16_+_31271274 | 0.12 |
ENST00000287497.8 ENST00000544665.3 |
ITGAM |
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chrX_+_15767971 | 0.12 |
ENST00000479740.1 ENST00000454127.2 |
CA5B |
carbonic anhydrase VB, mitochondrial |
| chr10_+_88414338 | 0.12 |
ENST00000241891.5 ENST00000443292.1 |
OPN4 |
opsin 4 |
| chr9_+_84603687 | 0.12 |
ENST00000344803.2 |
SPATA31D1 |
SPATA31 subfamily D, member 1 |
| chr12_-_39837192 | 0.12 |
ENST00000361961.3 ENST00000395670.3 |
KIF21A |
kinesin family member 21A |
| chr22_-_29784519 | 0.12 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr6_-_56507586 | 0.12 |
ENST00000439203.1 ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST |
dystonin |
| chr6_-_110011704 | 0.11 |
ENST00000448084.2 |
AK9 |
adenylate kinase 9 |
| chr12_+_112856690 | 0.11 |
ENST00000392597.1 ENST00000351677.2 |
PTPN11 |
protein tyrosine phosphatase, non-receptor type 11 |
| chr2_-_89521942 | 0.11 |
ENST00000482769.1 |
IGKV2-28 |
immunoglobulin kappa variable 2-28 |
| chr3_-_39322728 | 0.11 |
ENST00000541347.1 ENST00000412814.1 |
CX3CR1 |
chemokine (C-X3-C motif) receptor 1 |
| chr2_+_166326157 | 0.11 |
ENST00000421875.1 ENST00000314499.7 ENST00000409664.1 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
| chr2_-_187367356 | 0.11 |
ENST00000595956.1 |
AC018867.2 |
AC018867.2 |
| chr12_-_100660833 | 0.11 |
ENST00000551642.1 ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4 |
DEP domain containing 4 |
| chr3_-_21792838 | 0.11 |
ENST00000281523.2 |
ZNF385D |
zinc finger protein 385D |
| chr4_-_140544386 | 0.11 |
ENST00000561977.1 |
RP11-308D13.3 |
RP11-308D13.3 |
| chr17_+_7387677 | 0.10 |
ENST00000322644.6 |
POLR2A |
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr10_-_120101752 | 0.10 |
ENST00000369170.4 |
FAM204A |
family with sequence similarity 204, member A |
| chr20_-_1472029 | 0.10 |
ENST00000359801.3 |
SIRPB2 |
signal-regulatory protein beta 2 |
| chr15_-_31283618 | 0.10 |
ENST00000563714.1 |
MTMR10 |
myotubularin related protein 10 |
| chr14_+_22580233 | 0.10 |
ENST00000390454.2 |
TRAV25 |
T cell receptor alpha variable 25 |
| chr7_-_122339162 | 0.10 |
ENST00000340112.2 |
RNF133 |
ring finger protein 133 |
| chr3_+_137906353 | 0.10 |
ENST00000461822.1 ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr2_+_113816215 | 0.10 |
ENST00000346807.3 |
IL36RN |
interleukin 36 receptor antagonist |
| chr18_-_3845321 | 0.10 |
ENST00000539435.1 ENST00000400147.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr7_-_127255982 | 0.10 |
ENST00000338516.3 ENST00000378740.2 |
PAX4 |
paired box 4 |
| chr3_-_169587621 | 0.10 |
ENST00000523069.1 ENST00000316428.5 ENST00000264676.5 |
LRRC31 |
leucine rich repeat containing 31 |
| chr5_-_160973649 | 0.09 |
ENST00000393959.1 ENST00000517547.1 |
GABRB2 |
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr18_-_77711625 | 0.09 |
ENST00000357575.4 ENST00000590381.1 ENST00000397778.2 |
PQLC1 |
PQ loop repeat containing 1 |
| chr1_+_52082751 | 0.09 |
ENST00000447887.1 ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr3_+_156799587 | 0.09 |
ENST00000469196.1 |
RP11-6F2.5 |
RP11-6F2.5 |
| chr3_+_38347427 | 0.09 |
ENST00000273173.4 |
SLC22A14 |
solute carrier family 22, member 14 |
| chr7_-_14942283 | 0.09 |
ENST00000402815.1 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr19_+_45681997 | 0.09 |
ENST00000433642.2 |
BLOC1S3 |
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr18_+_56892724 | 0.09 |
ENST00000456142.3 ENST00000530323.1 |
GRP |
gastrin-releasing peptide |
| chr8_-_135522425 | 0.09 |
ENST00000521673.1 |
ZFAT |
zinc finger and AT hook domain containing |
| chr10_-_36813162 | 0.09 |
ENST00000440465.1 |
NAMPTL |
nicotinamide phosphoribosyltransferase-like |
| chr22_-_50523760 | 0.09 |
ENST00000395876.2 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr3_+_173116225 | 0.09 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr4_+_156680143 | 0.09 |
ENST00000505154.1 |
GUCY1B3 |
guanylate cyclase 1, soluble, beta 3 |
| chr11_+_125774362 | 0.09 |
ENST00000530414.1 ENST00000530129.2 |
DDX25 |
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr12_-_15103621 | 0.08 |
ENST00000536592.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr12_-_80084862 | 0.08 |
ENST00000328827.4 |
PAWR |
PRKC, apoptosis, WT1, regulator |
| chr4_-_100356551 | 0.08 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr1_+_235492300 | 0.08 |
ENST00000476121.1 ENST00000497327.1 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
| chr19_+_51728316 | 0.08 |
ENST00000436584.2 ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33 |
CD33 molecule |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.5 | 1.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.8 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.2 | 0.8 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.2 | 1.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 0.5 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 1.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 1.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.5 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.8 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 1.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.7 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.3 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.3 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 0.2 | GO:0006756 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.3 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.4 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.3 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.3 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 1.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.2 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 1.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.4 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 1.4 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.0 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.5 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 1.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.6 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.2 | 1.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.8 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.2 | 0.8 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.3 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.8 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.7 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.2 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 1.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 1.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |