Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for SOX9
Z-value: 0.12
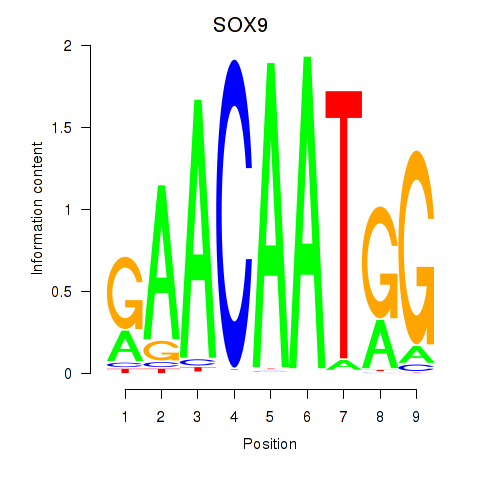
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SOX9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX9 | hg19_v2_chr17_+_70117153_70117174 | 0.53 | 1.8e-01 | Click! |
Activity profile of SOX9 motif
Sorted Z-values of SOX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_149051194 | 0.18 |
ENST00000470080.1 |
TM4SF18 |
transmembrane 4 L six family member 18 |
| chr17_-_49124230 | 0.16 |
ENST00000510283.1 ENST00000510855.1 |
SPAG9 |
sperm associated antigen 9 |
| chr3_-_65583561 | 0.15 |
ENST00000460329.2 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr2_+_30569506 | 0.14 |
ENST00000421976.2 |
AC109642.1 |
AC109642.1 |
| chr11_+_131781290 | 0.13 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
| chr7_-_131241361 | 0.13 |
ENST00000378555.3 ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL |
podocalyxin-like |
| chr6_-_128841503 | 0.12 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr1_+_64239657 | 0.12 |
ENST00000371080.1 ENST00000371079.1 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
| chr20_-_17662705 | 0.12 |
ENST00000455029.2 |
RRBP1 |
ribosome binding protein 1 |
| chr20_-_17662878 | 0.12 |
ENST00000377813.1 ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1 |
ribosome binding protein 1 |
| chr12_-_71003568 | 0.11 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr15_+_85923797 | 0.11 |
ENST00000559362.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr5_-_16936340 | 0.11 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr1_-_19426149 | 0.11 |
ENST00000429347.2 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
| chr3_-_8686479 | 0.11 |
ENST00000544814.1 ENST00000427408.1 |
SSUH2 |
ssu-2 homolog (C. elegans) |
| chr2_+_223536428 | 0.11 |
ENST00000446656.3 |
MOGAT1 |
monoacylglycerol O-acyltransferase 1 |
| chr10_-_129924611 | 0.10 |
ENST00000368654.3 |
MKI67 |
marker of proliferation Ki-67 |
| chr20_+_42086525 | 0.10 |
ENST00000244020.3 |
SRSF6 |
serine/arginine-rich splicing factor 6 |
| chr19_-_17375541 | 0.10 |
ENST00000252597.3 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr15_+_40886439 | 0.10 |
ENST00000532056.1 ENST00000399668.2 |
CASC5 |
cancer susceptibility candidate 5 |
| chr9_-_130517522 | 0.10 |
ENST00000373274.3 ENST00000420366.1 |
SH2D3C |
SH2 domain containing 3C |
| chr8_-_82395461 | 0.10 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chr21_+_26934165 | 0.09 |
ENST00000456917.1 |
MIR155HG |
MIR155 host gene (non-protein coding) |
| chr6_-_75915757 | 0.09 |
ENST00000322507.8 |
COL12A1 |
collagen, type XII, alpha 1 |
| chr8_+_98656693 | 0.09 |
ENST00000519934.1 |
MTDH |
metadherin |
| chr20_-_43150601 | 0.09 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr11_-_67141090 | 0.09 |
ENST00000312438.7 |
CLCF1 |
cardiotrophin-like cytokine factor 1 |
| chr3_+_9773409 | 0.09 |
ENST00000433861.2 ENST00000424362.1 ENST00000383829.2 ENST00000302054.3 ENST00000420291.1 |
BRPF1 |
bromodomain and PHD finger containing, 1 |
| chr5_-_146833485 | 0.09 |
ENST00000398514.3 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr7_-_92855762 | 0.09 |
ENST00000453812.2 ENST00000394468.2 |
HEPACAM2 |
HEPACAM family member 2 |
| chr6_+_114178512 | 0.09 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr12_-_45270151 | 0.09 |
ENST00000429094.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr1_+_43766642 | 0.09 |
ENST00000372476.3 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr3_-_149510553 | 0.08 |
ENST00000462519.2 ENST00000446160.1 ENST00000383050.3 |
ANKUB1 |
ankyrin repeat and ubiquitin domain containing 1 |
| chr17_+_26646121 | 0.08 |
ENST00000226230.6 |
TMEM97 |
transmembrane protein 97 |
| chr16_+_15528332 | 0.08 |
ENST00000566490.1 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr12_-_45270077 | 0.08 |
ENST00000551601.1 ENST00000549027.1 ENST00000452445.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr14_+_100070869 | 0.08 |
ENST00000502101.2 |
RP11-543C4.1 |
RP11-543C4.1 |
| chr1_-_182641367 | 0.08 |
ENST00000508450.1 |
RGS8 |
regulator of G-protein signaling 8 |
| chr3_-_46904918 | 0.08 |
ENST00000395869.1 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr1_-_93645818 | 0.08 |
ENST00000370280.1 ENST00000479918.1 |
TMED5 |
transmembrane emp24 protein transport domain containing 5 |
| chr6_+_56954867 | 0.08 |
ENST00000370708.4 ENST00000370702.1 |
ZNF451 |
zinc finger protein 451 |
| chrX_-_110654147 | 0.08 |
ENST00000358070.4 |
DCX |
doublecortin |
| chr12_-_122985494 | 0.08 |
ENST00000336229.4 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr5_+_74632993 | 0.08 |
ENST00000287936.4 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr15_+_85923856 | 0.08 |
ENST00000560302.1 ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr12_-_122985067 | 0.08 |
ENST00000540586.1 ENST00000543897.1 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr3_-_46904946 | 0.08 |
ENST00000292327.4 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr2_-_61765315 | 0.08 |
ENST00000406957.1 ENST00000401558.2 |
XPO1 |
exportin 1 (CRM1 homolog, yeast) |
| chr19_-_58514129 | 0.08 |
ENST00000552184.1 ENST00000546715.1 ENST00000536132.1 ENST00000547828.1 ENST00000547121.1 ENST00000551380.1 |
ZNF606 |
zinc finger protein 606 |
| chr19_-_17375527 | 0.08 |
ENST00000431146.2 ENST00000594190.1 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr5_+_74633036 | 0.08 |
ENST00000343975.5 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr7_+_138145076 | 0.08 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr3_+_39509163 | 0.08 |
ENST00000436143.2 ENST00000441980.2 ENST00000311042.6 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chr17_-_37764128 | 0.08 |
ENST00000302584.4 |
NEUROD2 |
neuronal differentiation 2 |
| chr7_-_111846435 | 0.08 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr15_+_40886199 | 0.08 |
ENST00000346991.5 ENST00000528975.1 ENST00000527044.1 |
CASC5 |
cancer susceptibility candidate 5 |
| chr9_-_35828576 | 0.07 |
ENST00000377984.2 ENST00000423537.2 ENST00000472182.1 |
FAM221B |
family with sequence similarity 221, member B |
| chr8_+_98656336 | 0.07 |
ENST00000336273.3 |
MTDH |
metadherin |
| chr19_+_58790314 | 0.07 |
ENST00000196548.5 ENST00000608843.1 |
ZNF8 ZNF8 |
Zinc finger protein 8 zinc finger protein 8 |
| chr12_-_111358372 | 0.07 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chr19_-_45004556 | 0.07 |
ENST00000587047.1 ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180 |
zinc finger protein 180 |
| chr1_+_185703513 | 0.07 |
ENST00000271588.4 ENST00000367492.2 |
HMCN1 |
hemicentin 1 |
| chr12_-_24103954 | 0.07 |
ENST00000441133.2 ENST00000545921.1 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr1_-_68698222 | 0.07 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr7_+_138145145 | 0.07 |
ENST00000415680.2 |
TRIM24 |
tripartite motif containing 24 |
| chr4_+_41614720 | 0.07 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr14_-_88459503 | 0.07 |
ENST00000393568.4 ENST00000261304.2 |
GALC |
galactosylceramidase |
| chr4_+_95128748 | 0.07 |
ENST00000359052.4 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr5_+_140602904 | 0.07 |
ENST00000515856.2 ENST00000239449.4 |
PCDHB14 |
protocadherin beta 14 |
| chr10_-_104953009 | 0.07 |
ENST00000470299.1 ENST00000343289.5 |
NT5C2 |
5'-nucleotidase, cytosolic II |
| chr11_-_69519410 | 0.07 |
ENST00000294312.3 |
FGF19 |
fibroblast growth factor 19 |
| chr19_-_51891209 | 0.07 |
ENST00000221973.3 ENST00000596399.1 |
LIM2 |
lens intrinsic membrane protein 2, 19kDa |
| chr10_+_70480963 | 0.07 |
ENST00000265872.6 ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
| chr5_+_40679584 | 0.07 |
ENST00000302472.3 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
| chr2_-_97405775 | 0.07 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr9_+_134000948 | 0.07 |
ENST00000359428.5 ENST00000411637.2 ENST00000451030.1 |
NUP214 |
nucleoporin 214kDa |
| chr14_-_53019211 | 0.07 |
ENST00000557374.1 ENST00000281741.4 |
TXNDC16 |
thioredoxin domain containing 16 |
| chr15_-_34502197 | 0.07 |
ENST00000557877.1 |
KATNBL1 |
katanin p80 subunit B-like 1 |
| chr1_+_93645314 | 0.07 |
ENST00000343253.7 |
CCDC18 |
coiled-coil domain containing 18 |
| chr6_-_53213780 | 0.07 |
ENST00000304434.6 ENST00000370918.4 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
| chr3_+_189507460 | 0.07 |
ENST00000434928.1 |
TP63 |
tumor protein p63 |
| chr22_+_38142235 | 0.07 |
ENST00000407319.2 ENST00000403663.2 ENST00000428075.1 |
TRIOBP |
TRIO and F-actin binding protein |
| chr10_+_73724123 | 0.07 |
ENST00000373115.4 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr16_+_3019552 | 0.07 |
ENST00000572687.1 |
PAQR4 |
progestin and adipoQ receptor family member IV |
| chr12_-_91546926 | 0.07 |
ENST00000550758.1 |
DCN |
decorin |
| chr2_-_70475701 | 0.06 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr7_+_99775366 | 0.06 |
ENST00000394018.2 ENST00000416412.1 |
STAG3 |
stromal antigen 3 |
| chr10_+_6244829 | 0.06 |
ENST00000317350.4 ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr3_+_39509070 | 0.06 |
ENST00000354668.4 ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chrM_+_4431 | 0.06 |
ENST00000361453.3 |
MT-ND2 |
mitochondrially encoded NADH dehydrogenase 2 |
| chr2_+_54683419 | 0.06 |
ENST00000356805.4 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr7_-_71868354 | 0.06 |
ENST00000412588.1 |
CALN1 |
calneuron 1 |
| chr1_-_40367530 | 0.06 |
ENST00000372816.2 ENST00000372815.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr7_-_121944491 | 0.06 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr2_-_200323414 | 0.06 |
ENST00000443023.1 |
SATB2 |
SATB homeobox 2 |
| chr15_+_90895471 | 0.06 |
ENST00000354377.3 ENST00000379090.5 |
ZNF774 |
zinc finger protein 774 |
| chr11_-_10829851 | 0.06 |
ENST00000532082.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr5_-_146833222 | 0.06 |
ENST00000534907.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr17_+_26646175 | 0.06 |
ENST00000583381.1 ENST00000582113.1 ENST00000582384.1 |
TMEM97 |
transmembrane protein 97 |
| chr12_-_18243119 | 0.06 |
ENST00000538724.1 ENST00000229002.2 |
RERGL |
RERG/RAS-like |
| chrX_-_53711064 | 0.06 |
ENST00000342160.3 ENST00000446750.1 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr5_-_141338627 | 0.06 |
ENST00000231484.3 |
PCDH12 |
protocadherin 12 |
| chr7_-_112635675 | 0.06 |
ENST00000447785.1 ENST00000451962.1 |
AC018464.3 |
AC018464.3 |
| chr1_-_156828810 | 0.06 |
ENST00000368195.3 |
INSRR |
insulin receptor-related receptor |
| chr9_-_73483958 | 0.06 |
ENST00000377101.1 ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr10_-_62332357 | 0.06 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr14_+_53019993 | 0.06 |
ENST00000542169.2 ENST00000555622.1 |
GPR137C |
G protein-coupled receptor 137C |
| chr1_+_180897269 | 0.06 |
ENST00000367587.1 |
KIAA1614 |
KIAA1614 |
| chr6_-_39282329 | 0.06 |
ENST00000373231.4 |
KCNK17 |
potassium channel, subfamily K, member 17 |
| chr20_-_40247133 | 0.06 |
ENST00000373233.3 ENST00000309279.7 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
| chr4_-_5894777 | 0.06 |
ENST00000324989.7 |
CRMP1 |
collapsin response mediator protein 1 |
| chr17_+_77751931 | 0.06 |
ENST00000310942.4 ENST00000269399.5 |
CBX2 |
chromobox homolog 2 |
| chr7_+_11013491 | 0.06 |
ENST00000403050.3 ENST00000445996.2 |
PHF14 |
PHD finger protein 14 |
| chrX_-_73512411 | 0.06 |
ENST00000602576.1 ENST00000429124.1 |
FTX |
FTX transcript, XIST regulator (non-protein coding) |
| chr20_-_45984401 | 0.06 |
ENST00000311275.7 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr16_-_89556942 | 0.06 |
ENST00000301030.4 |
ANKRD11 |
ankyrin repeat domain 11 |
| chr16_+_3068393 | 0.05 |
ENST00000573001.1 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
| chr1_-_243418621 | 0.05 |
ENST00000366544.1 ENST00000366543.1 |
CEP170 |
centrosomal protein 170kDa |
| chr9_-_21482312 | 0.05 |
ENST00000448696.3 |
IFNE |
interferon, epsilon |
| chr8_-_38326139 | 0.05 |
ENST00000335922.5 ENST00000532791.1 ENST00000397091.5 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr11_-_77531752 | 0.05 |
ENST00000440064.2 ENST00000528095.1 |
RSF1 |
remodeling and spacing factor 1 |
| chr2_-_70475730 | 0.05 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr11_+_67159416 | 0.05 |
ENST00000307980.2 ENST00000544620.1 |
RAD9A |
RAD9 homolog A (S. pombe) |
| chr16_+_3019246 | 0.05 |
ENST00000318782.8 ENST00000293978.8 |
PAQR4 |
progestin and adipoQ receptor family member IV |
| chr10_-_13570533 | 0.05 |
ENST00000396900.2 ENST00000396898.2 |
BEND7 |
BEN domain containing 7 |
| chr2_-_55277654 | 0.05 |
ENST00000337526.6 ENST00000317610.7 ENST00000357732.4 |
RTN4 |
reticulon 4 |
| chr8_-_70745575 | 0.05 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr16_-_67217844 | 0.05 |
ENST00000563902.1 ENST00000561621.1 ENST00000290881.7 |
KIAA0895L |
KIAA0895-like |
| chr3_-_112329110 | 0.05 |
ENST00000479368.1 |
CCDC80 |
coiled-coil domain containing 80 |
| chr7_+_100450328 | 0.05 |
ENST00000540482.1 ENST00000418037.1 ENST00000428758.1 ENST00000275729.3 ENST00000415287.1 ENST00000354161.3 ENST00000416675.1 |
SLC12A9 |
solute carrier family 12, member 9 |
| chr7_+_155090271 | 0.05 |
ENST00000476756.1 |
INSIG1 |
insulin induced gene 1 |
| chr4_-_41750922 | 0.05 |
ENST00000226382.2 |
PHOX2B |
paired-like homeobox 2b |
| chr11_-_8190534 | 0.05 |
ENST00000309737.6 ENST00000425599.2 ENST00000539720.1 ENST00000531450.1 ENST00000419822.2 ENST00000335425.7 ENST00000343202.4 |
RIC3 |
RIC3 acetylcholine receptor chaperone |
| chr2_-_55277692 | 0.05 |
ENST00000394611.2 |
RTN4 |
reticulon 4 |
| chr16_+_67261008 | 0.05 |
ENST00000304800.9 ENST00000563953.1 ENST00000565201.1 |
TMEM208 |
transmembrane protein 208 |
| chr4_+_95128996 | 0.05 |
ENST00000457823.2 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr16_-_73093597 | 0.05 |
ENST00000397992.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr7_-_38289173 | 0.05 |
ENST00000436911.2 |
TRGC2 |
T cell receptor gamma constant 2 |
| chr6_+_72596406 | 0.05 |
ENST00000491071.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr6_+_72596604 | 0.05 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr5_+_140739537 | 0.05 |
ENST00000522605.1 |
PCDHGB2 |
protocadherin gamma subfamily B, 2 |
| chrX_+_85403487 | 0.05 |
ENST00000373125.4 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chrX_-_38080077 | 0.05 |
ENST00000378533.3 ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX |
sushi-repeat containing protein, X-linked |
| chr22_+_31090793 | 0.05 |
ENST00000332585.6 ENST00000382310.3 ENST00000446658.2 |
OSBP2 |
oxysterol binding protein 2 |
| chr5_-_43313574 | 0.05 |
ENST00000325110.6 ENST00000433297.2 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr6_+_32132360 | 0.05 |
ENST00000333845.6 ENST00000395512.1 ENST00000432129.1 |
EGFL8 |
EGF-like-domain, multiple 8 |
| chr18_-_31802282 | 0.05 |
ENST00000535475.1 |
NOL4 |
nucleolar protein 4 |
| chr5_+_140213815 | 0.05 |
ENST00000525929.1 ENST00000378125.3 |
PCDHA7 |
protocadherin alpha 7 |
| chr10_-_128359074 | 0.05 |
ENST00000544758.1 |
C10orf90 |
chromosome 10 open reading frame 90 |
| chr7_+_120590803 | 0.05 |
ENST00000315870.5 ENST00000339121.5 ENST00000445699.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr14_+_85996507 | 0.05 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr18_+_42260861 | 0.05 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr16_-_15474904 | 0.05 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr12_-_11548496 | 0.05 |
ENST00000389362.4 ENST00000565533.1 ENST00000546254.1 |
PRB2 PRB1 |
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr3_-_149051444 | 0.05 |
ENST00000296059.2 |
TM4SF18 |
transmembrane 4 L six family member 18 |
| chr4_+_95129061 | 0.05 |
ENST00000354268.4 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_-_21503337 | 0.05 |
ENST00000400422.1 ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chrX_+_48380205 | 0.05 |
ENST00000446158.1 ENST00000414061.1 |
EBP |
emopamil binding protein (sterol isomerase) |
| chr5_+_15500280 | 0.05 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chrX_+_85403445 | 0.05 |
ENST00000373131.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr1_-_243418344 | 0.05 |
ENST00000366542.1 |
CEP170 |
centrosomal protein 170kDa |
| chr11_-_46867780 | 0.05 |
ENST00000529230.1 ENST00000415402.1 ENST00000312055.5 |
CKAP5 |
cytoskeleton associated protein 5 |
| chr20_-_45985172 | 0.05 |
ENST00000536340.1 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr2_-_55277436 | 0.05 |
ENST00000354474.6 |
RTN4 |
reticulon 4 |
| chr11_+_67007518 | 0.05 |
ENST00000530342.1 ENST00000308783.5 |
KDM2A |
lysine (K)-specific demethylase 2A |
| chr1_-_197115818 | 0.04 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr10_+_104678032 | 0.04 |
ENST00000369878.4 ENST00000369875.3 |
CNNM2 |
cyclin M2 |
| chr15_-_59041768 | 0.04 |
ENST00000402627.1 ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10 |
ADAM metallopeptidase domain 10 |
| chr2_-_55277512 | 0.04 |
ENST00000402434.2 |
RTN4 |
reticulon 4 |
| chr12_+_124997766 | 0.04 |
ENST00000543970.1 |
RP11-83B20.1 |
RP11-83B20.1 |
| chr10_+_102106829 | 0.04 |
ENST00000370355.2 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr2_-_55276320 | 0.04 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chr14_+_75230011 | 0.04 |
ENST00000552421.1 ENST00000325680.7 ENST00000238571.3 |
YLPM1 |
YLP motif containing 1 |
| chr14_+_85996471 | 0.04 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr19_+_45394477 | 0.04 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr11_+_1411503 | 0.04 |
ENST00000526678.1 |
BRSK2 |
BR serine/threonine kinase 2 |
| chr17_-_62308087 | 0.04 |
ENST00000583097.1 |
TEX2 |
testis expressed 2 |
| chr5_+_140729649 | 0.04 |
ENST00000523390.1 |
PCDHGB1 |
protocadherin gamma subfamily B, 1 |
| chr5_-_147162263 | 0.04 |
ENST00000333010.6 ENST00000265272.5 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
| chr20_+_3827459 | 0.04 |
ENST00000416600.2 ENST00000428216.2 |
MAVS |
mitochondrial antiviral signaling protein |
| chr2_+_102314161 | 0.04 |
ENST00000425019.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr8_-_38325219 | 0.04 |
ENST00000533668.1 ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr16_-_27561209 | 0.04 |
ENST00000356183.4 ENST00000561623.1 |
GTF3C1 |
general transcription factor IIIC, polypeptide 1, alpha 220kDa |
| chr5_+_140248518 | 0.04 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chr14_-_88459182 | 0.04 |
ENST00000544807.2 |
GALC |
galactosylceramidase |
| chr20_+_30795664 | 0.04 |
ENST00000375749.3 ENST00000375730.3 ENST00000539210.1 |
POFUT1 |
protein O-fucosyltransferase 1 |
| chrX_-_153599578 | 0.04 |
ENST00000360319.4 ENST00000344736.4 |
FLNA |
filamin A, alpha |
| chr8_-_21645804 | 0.04 |
ENST00000518077.1 ENST00000517892.1 |
GFRA2 |
GDNF family receptor alpha 2 |
| chr11_-_79151695 | 0.04 |
ENST00000278550.7 |
TENM4 |
teneurin transmembrane protein 4 |
| chr7_+_116312411 | 0.04 |
ENST00000456159.1 ENST00000397752.3 ENST00000318493.6 |
MET |
met proto-oncogene |
| chr1_-_26324534 | 0.04 |
ENST00000374284.1 ENST00000441420.1 ENST00000374282.3 |
PAFAH2 |
platelet-activating factor acetylhydrolase 2, 40kDa |
| chr15_-_42186248 | 0.04 |
ENST00000320955.6 |
SPTBN5 |
spectrin, beta, non-erythrocytic 5 |
| chr8_-_38326119 | 0.04 |
ENST00000356207.5 ENST00000326324.6 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chrX_+_78003204 | 0.04 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr2_-_158182410 | 0.04 |
ENST00000419116.2 ENST00000410096.1 |
ERMN |
ermin, ERM-like protein |
| chr1_+_78769549 | 0.04 |
ENST00000370758.1 |
PTGFR |
prostaglandin F receptor (FP) |
| chr3_-_48700310 | 0.04 |
ENST00000164024.4 ENST00000544264.1 |
CELSR3 |
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr3_+_49507559 | 0.04 |
ENST00000421560.1 ENST00000308775.2 ENST00000545947.1 ENST00000541308.1 ENST00000539901.1 ENST00000538711.1 ENST00000418588.1 |
DAG1 |
dystroglycan 1 (dystrophin-associated glycoprotein 1) |
| chr9_+_2622085 | 0.04 |
ENST00000382099.2 |
VLDLR |
very low density lipoprotein receptor |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.0 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.0 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.0 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.0 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |