Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for STAT4
Z-value: 0.74
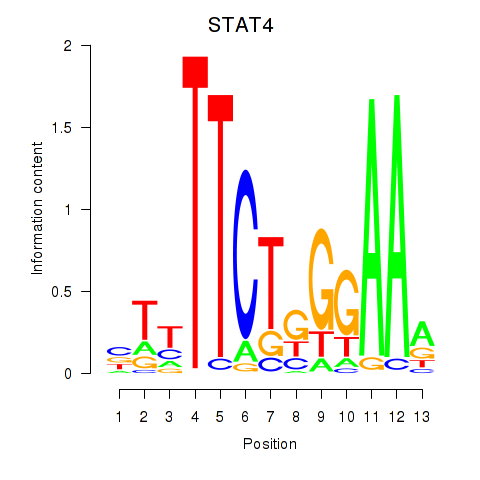
Transcription factors associated with STAT4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT4
|
ENSG00000138378.13 | STAT4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT4 | hg19_v2_chr2_-_192016316_192016325 | -0.14 | 7.4e-01 | Click! |
Activity profile of STAT4 motif
Sorted Z-values of STAT4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_120685608 | 0.93 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_-_111092873 | 0.87 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr2_-_190044480 | 0.85 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr14_-_30396948 | 0.75 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr5_-_111093081 | 0.73 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093340 | 0.72 |
ENST00000508870.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093167 | 0.66 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr5_-_111092930 | 0.62 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093406 | 0.61 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chr2_+_210444748 | 0.53 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr15_+_63340775 | 0.50 |
ENST00000559281.1 ENST00000317516.7 |
TPM1 |
tropomyosin 1 (alpha) |
| chr5_-_121413974 | 0.48 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr5_+_140753444 | 0.46 |
ENST00000517434.1 |
PCDHGA6 |
protocadherin gamma subfamily A, 6 |
| chr19_+_35225060 | 0.46 |
ENST00000599244.1 ENST00000392232.3 |
ZNF181 |
zinc finger protein 181 |
| chr15_+_63340858 | 0.43 |
ENST00000560615.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr11_+_117070904 | 0.43 |
ENST00000529792.1 |
TAGLN |
transgelin |
| chr15_+_63340734 | 0.40 |
ENST00000560959.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr12_+_53443963 | 0.38 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr6_+_32821924 | 0.38 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_-_202563414 | 0.35 |
ENST00000409474.3 ENST00000315506.7 ENST00000359962.5 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr8_-_27469196 | 0.34 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr1_-_144994909 | 0.34 |
ENST00000369347.4 ENST00000369354.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr6_+_36646435 | 0.31 |
ENST00000244741.5 ENST00000405375.1 ENST00000373711.2 |
CDKN1A |
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| chr3_-_156534754 | 0.31 |
ENST00000472943.1 ENST00000473352.1 |
LINC00886 |
long intergenic non-protein coding RNA 886 |
| chr19_-_44405941 | 0.31 |
ENST00000587128.1 |
RP11-15A1.3 |
RP11-15A1.3 |
| chr6_+_108977520 | 0.31 |
ENST00000540898.1 |
FOXO3 |
forkhead box O3 |
| chr1_+_202431859 | 0.30 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr15_-_52944231 | 0.29 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr5_+_137203465 | 0.28 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr1_-_109655355 | 0.28 |
ENST00000369945.3 |
C1orf194 |
chromosome 1 open reading frame 194 |
| chr6_+_84222220 | 0.27 |
ENST00000369700.3 |
PRSS35 |
protease, serine, 35 |
| chr3_-_114866084 | 0.27 |
ENST00000357258.3 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr2_-_202562774 | 0.27 |
ENST00000396886.3 ENST00000409143.1 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr15_+_63340647 | 0.27 |
ENST00000404484.4 |
TPM1 |
tropomyosin 1 (alpha) |
| chr19_+_10197463 | 0.26 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr14_+_24630465 | 0.26 |
ENST00000557894.1 ENST00000559284.1 ENST00000560275.1 |
IRF9 |
interferon regulatory factor 9 |
| chr8_-_27468842 | 0.25 |
ENST00000523500.1 |
CLU |
clusterin |
| chr5_-_127674883 | 0.25 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr19_-_44405623 | 0.25 |
ENST00000591815.1 |
RP11-15A1.3 |
RP11-15A1.3 |
| chr6_+_32811861 | 0.24 |
ENST00000453426.1 |
TAPSAR1 |
TAP1 and PSMB8 antisense RNA 1 |
| chrX_-_63005405 | 0.23 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr19_+_44617511 | 0.23 |
ENST00000262894.6 ENST00000588926.1 ENST00000592780.1 |
ZNF225 |
zinc finger protein 225 |
| chr19_-_58864848 | 0.22 |
ENST00000263100.3 |
A1BG |
alpha-1-B glycoprotein |
| chr16_-_67260691 | 0.22 |
ENST00000447579.1 ENST00000393992.1 ENST00000424285.1 |
LRRC29 |
leucine rich repeat containing 29 |
| chr2_-_175547571 | 0.22 |
ENST00000409415.3 ENST00000359761.3 ENST00000272746.5 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr19_+_35168567 | 0.21 |
ENST00000457781.2 ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302 |
zinc finger protein 302 |
| chrX_+_54834159 | 0.21 |
ENST00000375053.2 ENST00000347546.4 ENST00000375062.4 |
MAGED2 |
melanoma antigen family D, 2 |
| chr7_-_108168580 | 0.21 |
ENST00000453085.1 |
PNPLA8 |
patatin-like phospholipase domain containing 8 |
| chr9_-_95166841 | 0.21 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr9_-_95166884 | 0.21 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr9_-_116163400 | 0.20 |
ENST00000277315.5 ENST00000448137.1 ENST00000409155.3 |
ALAD |
aminolevulinate dehydratase |
| chr22_-_50699701 | 0.20 |
ENST00000395780.1 |
MAPK12 |
mitogen-activated protein kinase 12 |
| chr11_+_20044096 | 0.20 |
ENST00000533917.1 |
NAV2 |
neuron navigator 2 |
| chr19_-_44952635 | 0.20 |
ENST00000592308.1 ENST00000588931.1 ENST00000291187.4 |
ZNF229 |
zinc finger protein 229 |
| chr1_-_68299130 | 0.20 |
ENST00000370982.3 |
GNG12 |
guanine nucleotide binding protein (G protein), gamma 12 |
| chr11_+_6411670 | 0.19 |
ENST00000530395.1 ENST00000527275.1 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr7_+_55980331 | 0.19 |
ENST00000429591.2 |
ZNF713 |
zinc finger protein 713 |
| chr19_+_35168633 | 0.19 |
ENST00000505365.2 |
ZNF302 |
zinc finger protein 302 |
| chr16_+_4674787 | 0.19 |
ENST00000262370.7 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr6_+_46761118 | 0.18 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr11_-_89223883 | 0.18 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chrX_+_106045891 | 0.18 |
ENST00000357242.5 ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
| chr16_+_4674814 | 0.17 |
ENST00000415496.1 ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr18_+_6729698 | 0.17 |
ENST00000383472.4 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr11_+_65657875 | 0.17 |
ENST00000312579.2 |
CCDC85B |
coiled-coil domain containing 85B |
| chr11_-_89224139 | 0.17 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr8_+_38586068 | 0.16 |
ENST00000443286.2 ENST00000520340.1 ENST00000518415.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr6_-_32811771 | 0.16 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr19_-_50529193 | 0.16 |
ENST00000596445.1 ENST00000599538.1 |
VRK3 |
vaccinia related kinase 3 |
| chr19_+_44669280 | 0.15 |
ENST00000590089.1 ENST00000454662.2 |
ZNF226 |
zinc finger protein 226 |
| chr9_+_100069933 | 0.15 |
ENST00000529487.1 |
CCDC180 |
coiled-coil domain containing 180 |
| chr3_-_183735651 | 0.15 |
ENST00000427120.2 ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr10_-_104179682 | 0.15 |
ENST00000406432.1 |
PSD |
pleckstrin and Sec7 domain containing |
| chr12_+_26164645 | 0.14 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr6_+_35995552 | 0.14 |
ENST00000468133.1 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr14_+_55034599 | 0.14 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr1_-_109655377 | 0.14 |
ENST00000369948.3 |
C1orf194 |
chromosome 1 open reading frame 194 |
| chr1_-_144994840 | 0.14 |
ENST00000369351.3 ENST00000369349.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr14_-_25103472 | 0.14 |
ENST00000216341.4 ENST00000382542.1 ENST00000382540.1 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr19_+_35168547 | 0.14 |
ENST00000502743.1 ENST00000509528.1 ENST00000506901.1 |
ZNF302 |
zinc finger protein 302 |
| chr19_-_51875894 | 0.14 |
ENST00000600427.1 ENST00000595217.1 ENST00000221978.5 |
NKG7 |
natural killer cell group 7 sequence |
| chr6_+_35995488 | 0.14 |
ENST00000229795.3 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr1_+_155146318 | 0.14 |
ENST00000368385.4 ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46 |
tripartite motif containing 46 |
| chr9_-_85882145 | 0.13 |
ENST00000328788.1 |
FRMD3 |
FERM domain containing 3 |
| chr6_+_35995531 | 0.13 |
ENST00000229794.4 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr12_+_27396901 | 0.12 |
ENST00000541191.1 ENST00000389032.3 |
STK38L |
serine/threonine kinase 38 like |
| chr6_-_110011718 | 0.12 |
ENST00000532976.1 |
AK9 |
adenylate kinase 9 |
| chr19_+_56905024 | 0.12 |
ENST00000591172.1 ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1 |
ZNF582 antisense RNA 1 (head to head) |
| chr17_+_5323492 | 0.12 |
ENST00000405578.4 ENST00000574003.1 |
RPAIN |
RPA interacting protein |
| chr15_+_67458357 | 0.11 |
ENST00000537194.2 |
SMAD3 |
SMAD family member 3 |
| chr12_-_39299406 | 0.11 |
ENST00000331366.5 |
CPNE8 |
copine VIII |
| chr20_+_5731083 | 0.10 |
ENST00000445603.1 ENST00000442185.1 |
C20orf196 |
chromosome 20 open reading frame 196 |
| chr15_+_57998821 | 0.10 |
ENST00000299638.3 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr15_+_57998923 | 0.10 |
ENST00000380557.4 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr11_-_118436707 | 0.10 |
ENST00000264020.2 ENST00000264021.3 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
| chrX_+_135570046 | 0.10 |
ENST00000370648.3 |
BRS3 |
bombesin-like receptor 3 |
| chr21_+_38593701 | 0.10 |
ENST00000440629.1 |
AP001432.14 |
AP001432.14 |
| chr3_-_164913777 | 0.10 |
ENST00000475390.1 |
SLITRK3 |
SLIT and NTRK-like family, member 3 |
| chr10_+_102790980 | 0.10 |
ENST00000393459.1 ENST00000224807.5 |
SFXN3 |
sideroflexin 3 |
| chrX_-_107334750 | 0.10 |
ENST00000340200.5 ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chrX_-_70474910 | 0.10 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr18_+_6729725 | 0.10 |
ENST00000400091.2 ENST00000583410.1 ENST00000584387.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr19_+_37709076 | 0.10 |
ENST00000590503.1 ENST00000589413.1 |
ZNF383 |
zinc finger protein 383 |
| chrX_-_46187069 | 0.10 |
ENST00000446884.1 |
RP1-30G7.2 |
RP1-30G7.2 |
| chr3_+_122399444 | 0.10 |
ENST00000474629.2 |
PARP14 |
poly (ADP-ribose) polymerase family, member 14 |
| chr3_+_159570722 | 0.10 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr17_-_58603568 | 0.10 |
ENST00000083182.3 |
APPBP2 |
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chrX_-_107334790 | 0.09 |
ENST00000217958.3 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr20_+_5731027 | 0.09 |
ENST00000378979.4 ENST00000303142.6 |
C20orf196 |
chromosome 20 open reading frame 196 |
| chr1_+_9299895 | 0.09 |
ENST00000602477.1 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr4_-_140005443 | 0.09 |
ENST00000510408.1 ENST00000420916.2 ENST00000358635.3 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
| chr20_+_8112824 | 0.09 |
ENST00000378641.3 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr17_-_77924627 | 0.09 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr3_-_187455680 | 0.08 |
ENST00000438077.1 |
BCL6 |
B-cell CLL/lymphoma 6 |
| chr20_+_39657454 | 0.08 |
ENST00000361337.2 |
TOP1 |
topoisomerase (DNA) I |
| chr6_-_110011704 | 0.08 |
ENST00000448084.2 |
AK9 |
adenylate kinase 9 |
| chr12_-_11175219 | 0.08 |
ENST00000390673.2 |
TAS2R19 |
taste receptor, type 2, member 19 |
| chr1_-_8000872 | 0.08 |
ENST00000377507.3 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
| chr5_+_179921344 | 0.08 |
ENST00000261951.4 |
CNOT6 |
CCR4-NOT transcription complex, subunit 6 |
| chr14_+_65381079 | 0.08 |
ENST00000549115.1 ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1 FNTB CHURC1-FNTB |
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr16_-_67260901 | 0.08 |
ENST00000341546.3 ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29 AC040160.1 |
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr19_+_44669221 | 0.07 |
ENST00000590578.1 ENST00000589160.1 ENST00000337433.5 ENST00000586286.1 ENST00000585560.1 ENST00000586914.1 ENST00000588883.1 ENST00000413984.2 ENST00000588742.1 ENST00000300823.6 ENST00000585678.1 ENST00000586203.1 ENST00000590467.1 ENST00000588795.1 ENST00000588127.1 |
ZNF226 |
zinc finger protein 226 |
| chr10_+_104404644 | 0.07 |
ENST00000462202.2 |
TRIM8 |
tripartite motif containing 8 |
| chr1_+_161632937 | 0.07 |
ENST00000236937.9 ENST00000367961.4 ENST00000358671.5 |
FCGR2B |
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr1_+_161551101 | 0.07 |
ENST00000367962.4 ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B |
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr4_+_130017268 | 0.07 |
ENST00000425929.1 ENST00000508673.1 ENST00000508622.1 |
C4orf33 |
chromosome 4 open reading frame 33 |
| chr7_+_117120017 | 0.07 |
ENST00000003084.6 ENST00000454343.1 |
CFTR |
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr15_-_60690163 | 0.07 |
ENST00000558998.1 ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2 |
annexin A2 |
| chr4_-_187476721 | 0.07 |
ENST00000307161.5 |
MTNR1A |
melatonin receptor 1A |
| chr14_-_20881579 | 0.06 |
ENST00000556935.1 ENST00000262715.5 ENST00000556549.1 |
TEP1 |
telomerase-associated protein 1 |
| chr3_+_5020801 | 0.06 |
ENST00000256495.3 |
BHLHE40 |
basic helix-loop-helix family, member e40 |
| chr4_-_140477928 | 0.06 |
ENST00000274031.3 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
| chr1_-_231005310 | 0.06 |
ENST00000470540.1 |
C1orf198 |
chromosome 1 open reading frame 198 |
| chr5_+_137203541 | 0.06 |
ENST00000421631.2 |
MYOT |
myotilin |
| chr3_-_155461515 | 0.06 |
ENST00000399242.2 |
AC104472.1 |
CDNA FLJ26134 fis, clone TMS03713; Uncharacterized protein |
| chr19_+_39881951 | 0.06 |
ENST00000315588.5 ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29 |
mediator complex subunit 29 |
| chrX_+_100646190 | 0.05 |
ENST00000471855.1 |
RPL36A |
ribosomal protein L36a |
| chr5_+_137203557 | 0.05 |
ENST00000515645.1 |
MYOT |
myotilin |
| chr11_+_123986069 | 0.05 |
ENST00000456829.2 ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A |
von Willebrand factor A domain containing 5A |
| chr5_+_32585605 | 0.05 |
ENST00000265073.4 ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
| chr6_+_31540056 | 0.05 |
ENST00000418386.2 |
LTA |
lymphotoxin alpha |
| chr15_-_75660919 | 0.05 |
ENST00000569482.1 ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1 |
mannosidase, alpha, class 2C, member 1 |
| chr1_+_233086326 | 0.05 |
ENST00000366628.5 ENST00000366627.4 |
NTPCR |
nucleoside-triphosphatase, cancer-related |
| chr11_-_10590238 | 0.05 |
ENST00000256178.3 |
LYVE1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr3_-_108672742 | 0.05 |
ENST00000261047.3 |
GUCA1C |
guanylate cyclase activator 1C |
| chr19_-_59084647 | 0.05 |
ENST00000594234.1 ENST00000596039.1 |
MZF1 |
myeloid zinc finger 1 |
| chr1_+_182584314 | 0.04 |
ENST00000566297.1 |
RP11-317P15.4 |
RP11-317P15.4 |
| chr22_+_24551765 | 0.04 |
ENST00000337989.7 |
CABIN1 |
calcineurin binding protein 1 |
| chr3_+_155838337 | 0.04 |
ENST00000490337.1 ENST00000389636.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr11_-_133715394 | 0.04 |
ENST00000299140.3 ENST00000532889.1 |
SPATA19 |
spermatogenesis associated 19 |
| chr3_+_122296465 | 0.04 |
ENST00000483793.1 |
PARP15 |
poly (ADP-ribose) polymerase family, member 15 |
| chr3_-_15469006 | 0.04 |
ENST00000443029.1 ENST00000383790.3 ENST00000383789.5 |
METTL6 |
methyltransferase like 6 |
| chr1_+_17906970 | 0.04 |
ENST00000375415.1 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr20_-_21494654 | 0.03 |
ENST00000377142.4 |
NKX2-2 |
NK2 homeobox 2 |
| chr12_+_60083118 | 0.03 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr9_+_72658490 | 0.03 |
ENST00000377182.4 |
MAMDC2 |
MAM domain containing 2 |
| chr3_-_108672609 | 0.03 |
ENST00000393963.3 ENST00000471108.1 |
GUCA1C |
guanylate cyclase activator 1C |
| chr7_+_117120106 | 0.03 |
ENST00000426809.1 |
CFTR |
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr11_-_64490634 | 0.03 |
ENST00000377559.3 ENST00000265459.6 |
NRXN2 |
neurexin 2 |
| chr15_+_64680003 | 0.03 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr4_+_174292058 | 0.02 |
ENST00000296504.3 |
SAP30 |
Sin3A-associated protein, 30kDa |
| chr6_-_116833500 | 0.02 |
ENST00000356128.4 |
TRAPPC3L |
trafficking protein particle complex 3-like |
| chr7_-_115670804 | 0.02 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr15_-_60690932 | 0.02 |
ENST00000559818.1 |
ANXA2 |
annexin A2 |
| chr2_+_171785824 | 0.02 |
ENST00000452526.2 |
GORASP2 |
golgi reassembly stacking protein 2, 55kDa |
| chr19_-_39881777 | 0.02 |
ENST00000595564.1 ENST00000221265.3 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr1_-_161279749 | 0.02 |
ENST00000533357.1 ENST00000360451.6 ENST00000336559.4 |
MPZ |
myelin protein zero |
| chr4_+_158141899 | 0.02 |
ENST00000264426.9 ENST00000506284.1 |
GRIA2 |
glutamate receptor, ionotropic, AMPA 2 |
| chr7_-_115670792 | 0.02 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr13_-_45915221 | 0.02 |
ENST00000309246.5 ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1 |
tumor protein, translationally-controlled 1 |
| chr19_-_56904799 | 0.02 |
ENST00000589895.1 ENST00000589143.1 ENST00000301310.4 ENST00000586929.1 |
ZNF582 |
zinc finger protein 582 |
| chr14_-_100841670 | 0.02 |
ENST00000557297.1 ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS |
tryptophanyl-tRNA synthetase |
| chr4_-_155533787 | 0.02 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr6_+_134210243 | 0.02 |
ENST00000367882.4 |
TCF21 |
transcription factor 21 |
| chr7_+_31569365 | 0.01 |
ENST00000454513.1 ENST00000451887.2 |
CCDC129 |
coiled-coil domain containing 129 |
| chr12_-_91398796 | 0.01 |
ENST00000261172.3 ENST00000551767.1 |
EPYC |
epiphycan |
| chr19_+_57106647 | 0.01 |
ENST00000328070.6 |
ZNF71 |
zinc finger protein 71 |
| chr19_-_7812446 | 0.01 |
ENST00000394173.4 ENST00000301357.8 |
CD209 |
CD209 molecule |
| chr3_-_164875850 | 0.01 |
ENST00000472120.1 |
RP11-747D18.1 |
RP11-747D18.1 |
| chr3_-_196242233 | 0.01 |
ENST00000397537.2 |
SMCO1 |
single-pass membrane protein with coiled-coil domains 1 |
| chr1_+_68150744 | 0.01 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chr6_-_136610911 | 0.01 |
ENST00000530767.1 ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1 |
BCL2-associated transcription factor 1 |
| chr1_-_143767881 | 0.01 |
ENST00000419275.1 |
PPIAL4G |
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
| chr12_+_58138664 | 0.00 |
ENST00000257910.3 |
TSPAN31 |
tetraspanin 31 |
| chr4_+_158141843 | 0.00 |
ENST00000509417.1 ENST00000296526.7 |
GRIA2 |
glutamate receptor, ionotropic, AMPA 2 |
| chr8_-_123139423 | 0.00 |
ENST00000523792.1 |
RP11-398G24.2 |
RP11-398G24.2 |
| chr14_+_20811766 | 0.00 |
ENST00000250416.5 ENST00000527915.1 |
PARP2 |
poly (ADP-ribose) polymerase 2 |
| chr19_-_54663473 | 0.00 |
ENST00000222224.3 |
LENG1 |
leukocyte receptor cluster (LRC) member 1 |
| chr9_-_134585221 | 0.00 |
ENST00000372190.3 ENST00000427994.1 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr14_+_20811722 | 0.00 |
ENST00000429687.3 |
PARP2 |
poly (ADP-ribose) polymerase 2 |
| chr9_-_27297126 | 0.00 |
ENST00000380031.1 ENST00000537675.1 ENST00000380032.3 |
EQTN |
equatorin, sperm acrosome associated |
| chr16_+_58283814 | 0.00 |
ENST00000443128.2 ENST00000219299.4 |
CCDC113 |
coiled-coil domain containing 113 |
| chrX_+_144908928 | 0.00 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr2_+_231921574 | 0.00 |
ENST00000308696.6 ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.6 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 1.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.2 | GO:0006756 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.2 | GO:0070541 | response to platinum ion(GO:0070541) cellular response to lead ion(GO:0071284) |
| 0.1 | 0.3 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0043317 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.3 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0099612 | protein localization to axon(GO:0099612) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 1.6 | GO:0032059 | muscle thin filament tropomyosin(GO:0005862) bleb(GO:0032059) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.6 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |