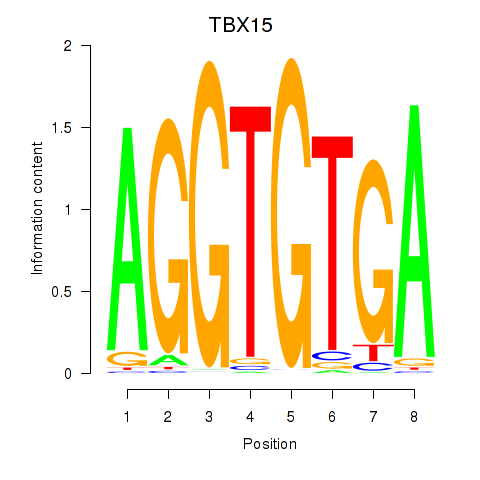
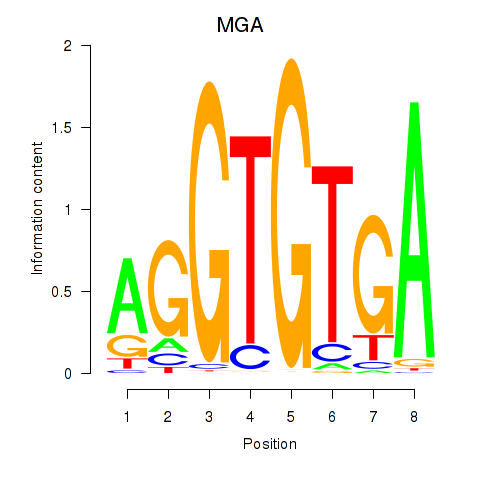
Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for TBX15_MGA
Z-value: 0.28
Transcription factors associated with TBX15_MGA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX15
|
ENSG00000092607.9 | TBX15 |
|
MGA
|
ENSG00000174197.12 | MGA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX15 | hg19_v2_chr1_-_119532127_119532179, hg19_v2_chr1_-_119530428_119530572 | -0.70 | 5.3e-02 | Click! |
| MGA | hg19_v2_chr15_+_41913690_41913753 | -0.41 | 3.1e-01 | Click! |
Activity profile of TBX15_MGA motif
Sorted Z-values of TBX15_MGA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX15_MGA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_22565101 | 0.27 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr14_+_75746781 | 0.22 |
ENST00000555347.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr1_+_33219592 | 0.22 |
ENST00000373481.3 |
KIAA1522 |
KIAA1522 |
| chr19_+_39279838 | 0.21 |
ENST00000314980.4 |
LGALS7B |
lectin, galactoside-binding, soluble, 7B |
| chr15_+_90319557 | 0.19 |
ENST00000341735.3 |
MESP2 |
mesoderm posterior 2 homolog (mouse) |
| chr10_+_94451574 | 0.18 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr19_-_46418033 | 0.17 |
ENST00000341294.2 |
NANOS2 |
nanos homolog 2 (Drosophila) |
| chr11_+_120081475 | 0.17 |
ENST00000328965.4 |
OAF |
OAF homolog (Drosophila) |
| chr1_-_205290865 | 0.16 |
ENST00000367157.3 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
| chr22_-_20231207 | 0.16 |
ENST00000425986.1 |
RTN4R |
reticulon 4 receptor |
| chr11_+_117947724 | 0.15 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr20_+_58630972 | 0.15 |
ENST00000313426.1 |
C20orf197 |
chromosome 20 open reading frame 197 |
| chr12_-_26278030 | 0.15 |
ENST00000242728.4 |
BHLHE41 |
basic helix-loop-helix family, member e41 |
| chr12_+_58148842 | 0.15 |
ENST00000266643.5 |
MARCH9 |
membrane-associated ring finger (C3HC4) 9 |
| chr4_-_170924888 | 0.15 |
ENST00000502832.1 ENST00000393704.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr6_-_13487784 | 0.15 |
ENST00000379287.3 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chr8_+_103563792 | 0.15 |
ENST00000285402.3 |
ODF1 |
outer dense fiber of sperm tails 1 |
| chr12_-_8815215 | 0.14 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr11_+_117947782 | 0.14 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr12_-_8815299 | 0.14 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr4_-_73434498 | 0.13 |
ENST00000286657.4 |
ADAMTS3 |
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr19_-_36001286 | 0.13 |
ENST00000602679.1 ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN |
dermokine |
| chr6_-_40555176 | 0.13 |
ENST00000338305.6 |
LRFN2 |
leucine rich repeat and fibronectin type III domain containing 2 |
| chrX_+_16804544 | 0.13 |
ENST00000380122.5 ENST00000398155.4 |
TXLNG |
taxilin gamma |
| chr8_+_123793633 | 0.12 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr7_-_105319536 | 0.12 |
ENST00000477775.1 |
ATXN7L1 |
ataxin 7-like 1 |
| chr8_+_102504651 | 0.12 |
ENST00000251808.3 ENST00000521085.1 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
| chr1_-_113258090 | 0.12 |
ENST00000309276.6 |
PPM1J |
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr11_-_119993979 | 0.12 |
ENST00000524816.3 ENST00000525327.1 |
TRIM29 |
tripartite motif containing 29 |
| chr1_-_113257905 | 0.12 |
ENST00000464951.1 |
PPM1J |
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr17_-_47841485 | 0.12 |
ENST00000506156.1 ENST00000240364.2 |
FAM117A |
family with sequence similarity 117, member A |
| chr12_+_56473939 | 0.12 |
ENST00000450146.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr17_-_46657473 | 0.12 |
ENST00000332503.5 |
HOXB4 |
homeobox B4 |
| chr6_+_36646435 | 0.12 |
ENST00000244741.5 ENST00000405375.1 ENST00000373711.2 |
CDKN1A |
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| chr7_-_76255444 | 0.11 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr9_-_114090713 | 0.11 |
ENST00000302681.1 ENST00000374428.1 |
OR2K2 |
olfactory receptor, family 2, subfamily K, member 2 |
| chr18_-_47376197 | 0.11 |
ENST00000592688.1 |
MYO5B |
myosin VB |
| chr4_+_57371509 | 0.11 |
ENST00000360096.2 |
ARL9 |
ADP-ribosylation factor-like 9 |
| chr12_-_8815404 | 0.11 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr9_+_102584128 | 0.10 |
ENST00000338488.4 ENST00000395097.2 |
NR4A3 |
nuclear receptor subfamily 4, group A, member 3 |
| chr10_+_47746929 | 0.10 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr1_-_153066998 | 0.10 |
ENST00000368750.3 |
SPRR2E |
small proline-rich protein 2E |
| chr1_-_25256368 | 0.10 |
ENST00000308873.6 |
RUNX3 |
runt-related transcription factor 3 |
| chr9_-_136024721 | 0.10 |
ENST00000393160.3 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
| chr19_-_36001113 | 0.10 |
ENST00000434389.1 |
DMKN |
dermokine |
| chr6_+_7541808 | 0.10 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr1_-_111174054 | 0.10 |
ENST00000369770.3 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr1_-_12677714 | 0.09 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr14_-_24036943 | 0.09 |
ENST00000556843.1 ENST00000397120.3 ENST00000557189.1 |
AP1G2 |
adaptor-related protein complex 1, gamma 2 subunit |
| chr3_+_142315225 | 0.09 |
ENST00000457734.2 ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1 |
plastin 1 |
| chr2_-_31637560 | 0.09 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr17_-_40428359 | 0.09 |
ENST00000293328.3 |
STAT5B |
signal transducer and activator of transcription 5B |
| chr3_-_48471454 | 0.09 |
ENST00000296440.6 ENST00000448774.2 |
PLXNB1 |
plexin B1 |
| chr3_+_10857885 | 0.09 |
ENST00000254488.2 ENST00000454147.1 |
SLC6A11 |
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr10_-_99531709 | 0.09 |
ENST00000266066.3 |
SFRP5 |
secreted frizzled-related protein 5 |
| chr11_-_119599794 | 0.09 |
ENST00000264025.3 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr11_-_119993734 | 0.09 |
ENST00000533302.1 |
TRIM29 |
tripartite motif containing 29 |
| chr17_-_34122596 | 0.08 |
ENST00000250144.8 |
MMP28 |
matrix metallopeptidase 28 |
| chr6_+_7541845 | 0.08 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr5_-_78809950 | 0.08 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chrX_-_128788914 | 0.08 |
ENST00000429967.1 ENST00000307484.6 |
APLN |
apelin |
| chr7_-_44365216 | 0.08 |
ENST00000358707.3 ENST00000457475.1 ENST00000440254.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr1_-_156786634 | 0.08 |
ENST00000392306.2 ENST00000368199.3 |
SH2D2A |
SH2 domain containing 2A |
| chr1_-_156786530 | 0.08 |
ENST00000368198.3 |
SH2D2A |
SH2 domain containing 2A |
| chr14_+_67291158 | 0.08 |
ENST00000555456.1 |
GPHN |
gephyrin |
| chr12_+_50451331 | 0.08 |
ENST00000228468.4 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chr1_+_26737253 | 0.08 |
ENST00000326279.6 |
LIN28A |
lin-28 homolog A (C. elegans) |
| chr1_-_153029980 | 0.08 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr11_-_7818520 | 0.08 |
ENST00000329434.2 |
OR5P2 |
olfactory receptor, family 5, subfamily P, member 2 |
| chr1_-_205744205 | 0.08 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr2_+_202937972 | 0.08 |
ENST00000541917.1 ENST00000295844.3 |
AC079354.1 |
uncharacterized protein KIAA2012 |
| chr9_+_139847347 | 0.08 |
ENST00000371632.3 |
LCN12 |
lipocalin 12 |
| chr11_+_73358594 | 0.08 |
ENST00000227214.6 ENST00000398494.4 ENST00000543085.1 |
PLEKHB1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr2_-_216003127 | 0.08 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr17_+_30771279 | 0.07 |
ENST00000261712.3 ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr20_+_62492566 | 0.07 |
ENST00000369916.3 |
ABHD16B |
abhydrolase domain containing 16B |
| chr22_-_42342733 | 0.07 |
ENST00000402420.1 |
CENPM |
centromere protein M |
| chr13_-_99667960 | 0.07 |
ENST00000448493.2 |
DOCK9 |
dedicator of cytokinesis 9 |
| chr7_+_140373619 | 0.07 |
ENST00000483369.1 |
ADCK2 |
aarF domain containing kinase 2 |
| chr1_+_15736359 | 0.07 |
ENST00000375980.4 |
EFHD2 |
EF-hand domain family, member D2 |
| chr17_-_46682321 | 0.07 |
ENST00000225648.3 ENST00000484302.2 |
HOXB6 |
homeobox B6 |
| chr18_+_47088401 | 0.07 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr7_+_143013198 | 0.07 |
ENST00000343257.2 |
CLCN1 |
chloride channel, voltage-sensitive 1 |
| chr7_-_27135591 | 0.07 |
ENST00000343060.4 ENST00000355633.5 |
HOXA1 |
homeobox A1 |
| chr7_-_44179972 | 0.07 |
ENST00000446581.1 |
MYL7 |
myosin, light chain 7, regulatory |
| chr21_-_31869451 | 0.07 |
ENST00000334058.2 |
KRTAP19-4 |
keratin associated protein 19-4 |
| chr11_-_85430204 | 0.07 |
ENST00000389958.3 ENST00000527794.1 |
SYTL2 |
synaptotagmin-like 2 |
| chr5_+_68788594 | 0.07 |
ENST00000396442.2 ENST00000380766.2 |
OCLN |
occludin |
| chr11_-_85430163 | 0.07 |
ENST00000529581.1 ENST00000533577.1 |
SYTL2 |
synaptotagmin-like 2 |
| chr2_+_176972000 | 0.07 |
ENST00000249504.5 |
HOXD11 |
homeobox D11 |
| chr15_+_40861487 | 0.07 |
ENST00000315616.7 ENST00000559271.1 |
RPUSD2 |
RNA pseudouridylate synthase domain containing 2 |
| chr21_+_30502806 | 0.07 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr1_-_27701307 | 0.07 |
ENST00000270879.4 ENST00000354982.2 |
FCN3 |
ficolin (collagen/fibrinogen domain containing) 3 |
| chr8_-_99954788 | 0.07 |
ENST00000523601.1 |
STK3 |
serine/threonine kinase 3 |
| chr2_-_113993020 | 0.06 |
ENST00000465084.1 |
PAX8 |
paired box 8 |
| chr1_+_26737292 | 0.06 |
ENST00000254231.4 |
LIN28A |
lin-28 homolog A (C. elegans) |
| chrX_-_32173579 | 0.06 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr6_-_86099898 | 0.06 |
ENST00000455071.1 |
RP11-30P6.6 |
RP11-30P6.6 |
| chr11_+_19138670 | 0.06 |
ENST00000446113.2 ENST00000399351.3 |
ZDHHC13 |
zinc finger, DHHC-type containing 13 |
| chr17_+_34431212 | 0.06 |
ENST00000394495.1 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr12_-_25801478 | 0.06 |
ENST00000540106.1 ENST00000445693.1 ENST00000545543.1 ENST00000542224.1 |
IFLTD1 |
intermediate filament tail domain containing 1 |
| chr1_+_93544791 | 0.06 |
ENST00000545708.1 ENST00000540243.1 ENST00000370298.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr20_-_3644046 | 0.06 |
ENST00000290417.2 ENST00000319242.3 |
GFRA4 |
GDNF family receptor alpha 4 |
| chr10_-_15413035 | 0.06 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chr3_-_93747425 | 0.06 |
ENST00000315099.2 |
STX19 |
syntaxin 19 |
| chr6_+_30294612 | 0.06 |
ENST00000440271.1 ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39 |
tripartite motif containing 39 |
| chr3_+_167453493 | 0.06 |
ENST00000295777.5 ENST00000472747.2 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr17_-_74533734 | 0.06 |
ENST00000589342.1 |
CYGB |
cytoglobin |
| chr12_-_8815477 | 0.06 |
ENST00000433590.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr3_-_52443799 | 0.06 |
ENST00000470173.1 ENST00000296288.5 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr12_+_22778291 | 0.06 |
ENST00000545979.1 |
ETNK1 |
ethanolamine kinase 1 |
| chr17_-_8021710 | 0.06 |
ENST00000380149.1 ENST00000448843.2 |
ALOXE3 |
arachidonate lipoxygenase 3 |
| chr7_+_76090993 | 0.06 |
ENST00000425780.1 ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2 |
deltex homolog 2 (Drosophila) |
| chr12_+_50451462 | 0.06 |
ENST00000447966.2 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chr9_+_136399929 | 0.06 |
ENST00000393060.1 |
ADAMTSL2 |
ADAMTS-like 2 |
| chr20_+_825275 | 0.05 |
ENST00000541082.1 |
FAM110A |
family with sequence similarity 110, member A |
| chr4_+_102268904 | 0.05 |
ENST00000527564.1 ENST00000529296.1 |
AP001816.1 |
Uncharacterized protein |
| chr1_+_158325684 | 0.05 |
ENST00000368162.2 |
CD1E |
CD1e molecule |
| chr5_-_49737184 | 0.05 |
ENST00000508934.1 ENST00000303221.5 |
EMB |
embigin |
| chrX_+_123095155 | 0.05 |
ENST00000371160.1 ENST00000435103.1 |
STAG2 |
stromal antigen 2 |
| chr19_+_13135731 | 0.05 |
ENST00000587260.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr17_+_34430980 | 0.05 |
ENST00000250151.4 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr20_+_2517253 | 0.05 |
ENST00000358864.1 |
TMC2 |
transmembrane channel-like 2 |
| chr5_-_9630463 | 0.05 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr1_+_155099927 | 0.05 |
ENST00000368407.3 |
EFNA1 |
ephrin-A1 |
| chr3_+_52811596 | 0.05 |
ENST00000542827.1 ENST00000273283.2 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr16_-_4395905 | 0.05 |
ENST00000571941.1 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chrX_+_56259316 | 0.05 |
ENST00000468660.1 |
KLF8 |
Kruppel-like factor 8 |
| chr18_-_45663666 | 0.05 |
ENST00000535628.2 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
| chr19_+_1077393 | 0.05 |
ENST00000590577.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr11_-_85430088 | 0.05 |
ENST00000533057.1 ENST00000533892.1 |
SYTL2 |
synaptotagmin-like 2 |
| chrX_+_105937068 | 0.05 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chrX_+_123095546 | 0.05 |
ENST00000371157.3 ENST00000371145.3 ENST00000371144.3 |
STAG2 |
stromal antigen 2 |
| chr6_+_30295036 | 0.05 |
ENST00000376659.5 ENST00000428555.1 |
TRIM39 |
tripartite motif containing 39 |
| chr7_-_150675372 | 0.05 |
ENST00000262186.5 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr15_-_88799384 | 0.05 |
ENST00000540489.2 ENST00000557856.1 ENST00000558676.1 |
NTRK3 |
neurotrophic tyrosine kinase, receptor, type 3 |
| chr14_+_22966642 | 0.05 |
ENST00000390496.1 |
TRAJ41 |
T cell receptor alpha joining 41 |
| chr2_-_208030647 | 0.05 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr5_-_34043310 | 0.05 |
ENST00000231338.7 |
C1QTNF3 |
C1q and tumor necrosis factor related protein 3 |
| chr17_-_40333099 | 0.05 |
ENST00000607371.1 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr8_-_130799134 | 0.05 |
ENST00000276708.4 |
GSDMC |
gasdermin C |
| chr19_-_42721819 | 0.05 |
ENST00000336034.4 ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2 |
death effector domain containing 2 |
| chr3_+_124223586 | 0.05 |
ENST00000393496.1 |
KALRN |
kalirin, RhoGEF kinase |
| chr22_-_31688381 | 0.05 |
ENST00000487265.2 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr12_-_95942613 | 0.05 |
ENST00000393091.2 |
USP44 |
ubiquitin specific peptidase 44 |
| chr16_-_81110683 | 0.05 |
ENST00000565253.1 ENST00000378611.4 ENST00000299578.5 |
C16orf46 |
chromosome 16 open reading frame 46 |
| chr12_+_18891045 | 0.05 |
ENST00000317658.3 |
CAPZA3 |
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr4_+_144434584 | 0.05 |
ENST00000283131.3 |
SMARCA5 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr11_-_44972476 | 0.05 |
ENST00000527685.1 ENST00000308212.5 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr1_-_153044083 | 0.05 |
ENST00000341611.2 |
SPRR2B |
small proline-rich protein 2B |
| chr1_+_182758900 | 0.05 |
ENST00000367555.1 ENST00000367554.3 |
NPL |
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
| chr1_-_226129083 | 0.05 |
ENST00000420304.2 |
LEFTY2 |
left-right determination factor 2 |
| chr1_+_155100342 | 0.04 |
ENST00000368406.2 |
EFNA1 |
ephrin-A1 |
| chr17_+_35849937 | 0.04 |
ENST00000394389.4 |
DUSP14 |
dual specificity phosphatase 14 |
| chr20_-_1309809 | 0.04 |
ENST00000360779.3 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chrX_-_48693955 | 0.04 |
ENST00000218230.5 |
PCSK1N |
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr6_-_13487825 | 0.04 |
ENST00000603223.1 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chr4_-_186456766 | 0.04 |
ENST00000284771.6 |
PDLIM3 |
PDZ and LIM domain 3 |
| chrX_+_122318113 | 0.04 |
ENST00000371264.3 |
GRIA3 |
glutamate receptor, ionotropic, AMPA 3 |
| chrX_+_70443050 | 0.04 |
ENST00000361726.6 |
GJB1 |
gap junction protein, beta 1, 32kDa |
| chr18_+_60382672 | 0.04 |
ENST00000400316.4 ENST00000262719.5 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
| chr17_-_10452929 | 0.04 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr4_+_154178520 | 0.04 |
ENST00000433687.1 |
TRIM2 |
tripartite motif containing 2 |
| chr7_-_71801980 | 0.04 |
ENST00000329008.5 |
CALN1 |
calneuron 1 |
| chr19_-_51875894 | 0.04 |
ENST00000600427.1 ENST00000595217.1 ENST00000221978.5 |
NKG7 |
natural killer cell group 7 sequence |
| chrX_-_11445856 | 0.04 |
ENST00000380736.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr17_-_7232585 | 0.04 |
ENST00000571887.1 ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4 |
neuralized E3 ubiquitin protein ligase 4 |
| chr16_-_67965756 | 0.04 |
ENST00000571044.1 ENST00000571605.1 |
CTRL |
chymotrypsin-like |
| chr4_-_186456652 | 0.04 |
ENST00000284767.5 ENST00000284770.5 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr4_-_152682129 | 0.04 |
ENST00000512306.1 ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112 |
PET112 homolog (yeast) |
| chr19_-_11689752 | 0.04 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr3_-_109056419 | 0.04 |
ENST00000335658.6 |
DPPA4 |
developmental pluripotency associated 4 |
| chr5_+_113697983 | 0.04 |
ENST00000264773.3 |
KCNN2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr5_+_161494521 | 0.04 |
ENST00000356592.3 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr3_-_39196049 | 0.04 |
ENST00000514182.1 |
CSRNP1 |
cysteine-serine-rich nuclear protein 1 |
| chr1_+_92417716 | 0.04 |
ENST00000402388.1 |
BRDT |
bromodomain, testis-specific |
| chr1_+_17906970 | 0.04 |
ENST00000375415.1 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr5_+_133861339 | 0.04 |
ENST00000282605.4 ENST00000361895.2 ENST00000402835.1 |
PHF15 |
jade family PHD finger 2 |
| chr7_-_143956815 | 0.04 |
ENST00000493325.1 |
OR2A7 |
olfactory receptor, family 2, subfamily A, member 7 |
| chr16_-_67450325 | 0.04 |
ENST00000348579.2 |
ZDHHC1 |
zinc finger, DHHC-type containing 1 |
| chrX_+_123480194 | 0.04 |
ENST00000371139.4 |
SH2D1A |
SH2 domain containing 1A |
| chr9_-_73483958 | 0.04 |
ENST00000377101.1 ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr19_-_46974664 | 0.04 |
ENST00000438932.2 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr4_+_41540160 | 0.04 |
ENST00000503057.1 ENST00000511496.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr2_+_11696464 | 0.04 |
ENST00000234142.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr9_+_125281420 | 0.04 |
ENST00000340750.1 |
OR1J4 |
olfactory receptor, family 1, subfamily J, member 4 |
| chr4_-_140222358 | 0.04 |
ENST00000505036.1 ENST00000544855.1 ENST00000539002.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr19_-_46974741 | 0.04 |
ENST00000313683.10 ENST00000602246.1 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr1_+_95582881 | 0.04 |
ENST00000370203.4 ENST00000456991.1 |
TMEM56 |
transmembrane protein 56 |
| chr12_-_49351303 | 0.04 |
ENST00000256682.4 |
ARF3 |
ADP-ribosylation factor 3 |
| chr20_+_816695 | 0.04 |
ENST00000246100.3 |
FAM110A |
family with sequence similarity 110, member A |
| chr6_+_130339710 | 0.04 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr12_-_49351228 | 0.04 |
ENST00000541959.1 ENST00000447318.2 |
ARF3 |
ADP-ribosylation factor 3 |
| chr2_+_74881355 | 0.04 |
ENST00000357877.2 |
SEMA4F |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr8_-_82395461 | 0.04 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chrX_+_123480375 | 0.04 |
ENST00000360027.4 |
SH2D1A |
SH2 domain containing 1A |
| chr2_-_160472952 | 0.04 |
ENST00000541068.2 ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr12_+_21525818 | 0.03 |
ENST00000240652.3 ENST00000542023.1 ENST00000537593.1 |
IAPP |
islet amyloid polypeptide |
| chr11_-_7847519 | 0.03 |
ENST00000328375.1 |
OR5P3 |
olfactory receptor, family 5, subfamily P, member 3 |
| chr22_-_41215291 | 0.03 |
ENST00000542412.1 ENST00000544408.1 |
SLC25A17 |
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.2 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1900205 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.0 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.0 | GO:0043132 | coenzyme A transport(GO:0015880) FAD transport(GO:0015883) coenzyme A transmembrane transport(GO:0035349) FAD transmembrane transport(GO:0035350) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.0 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.0 | GO:0060620 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.0 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.0 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.0 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) FAD transmembrane transporter activity(GO:0015230) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |