Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for TBX5
Z-value: 0.82
Transcription factors associated with TBX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
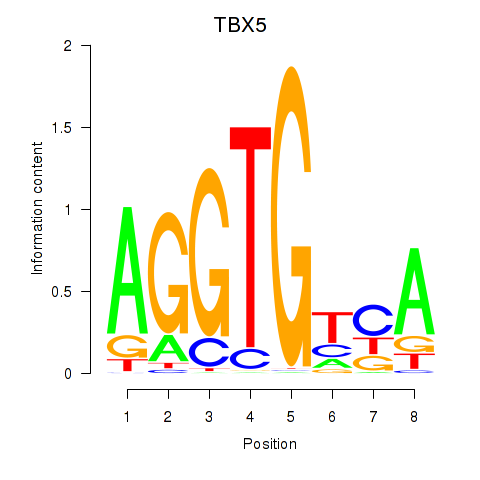
TBX5
|
ENSG00000089225.15 | TBX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX5 | hg19_v2_chr12_-_114841703_114841726 | 0.50 | 2.1e-01 | Click! |
Activity profile of TBX5 motif
Sorted Z-values of TBX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_117947724 | 1.26 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr11_+_117947782 | 1.15 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr12_-_52887034 | 0.87 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr1_+_101702417 | 0.68 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr22_-_20231207 | 0.67 |
ENST00000425986.1 |
RTN4R |
reticulon 4 receptor |
| chr6_+_150464155 | 0.67 |
ENST00000361131.4 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr16_-_31147020 | 0.59 |
ENST00000568261.1 ENST00000567797.1 ENST00000317508.6 |
PRSS8 |
protease, serine, 8 |
| chr1_-_205290865 | 0.57 |
ENST00000367157.3 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
| chr3_-_133748913 | 0.56 |
ENST00000310926.4 |
SLCO2A1 |
solute carrier organic anion transporter family, member 2A1 |
| chr2_+_68961934 | 0.53 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr12_+_107712173 | 0.53 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr2_+_68961905 | 0.53 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr8_+_120220561 | 0.50 |
ENST00000276681.6 |
MAL2 |
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr2_-_216003127 | 0.48 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr1_-_12677714 | 0.47 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr18_+_33877654 | 0.47 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr18_-_52969844 | 0.47 |
ENST00000561831.3 |
TCF4 |
transcription factor 4 |
| chr20_+_58630972 | 0.44 |
ENST00000313426.1 |
C20orf197 |
chromosome 20 open reading frame 197 |
| chr14_+_75746781 | 0.44 |
ENST00000555347.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr1_+_1981890 | 0.44 |
ENST00000378567.3 ENST00000468310.1 |
PRKCZ |
protein kinase C, zeta |
| chr15_+_40532058 | 0.43 |
ENST00000260404.4 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr12_-_28124903 | 0.43 |
ENST00000395872.1 ENST00000354417.3 ENST00000201015.4 |
PTHLH |
parathyroid hormone-like hormone |
| chr8_+_32405785 | 0.42 |
ENST00000287842.3 |
NRG1 |
neuregulin 1 |
| chr5_+_66124590 | 0.41 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr19_+_38755042 | 0.41 |
ENST00000301244.7 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr19_-_16045220 | 0.41 |
ENST00000326742.8 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr8_+_32405728 | 0.40 |
ENST00000523079.1 ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1 |
neuregulin 1 |
| chr8_-_21988558 | 0.39 |
ENST00000312841.8 |
HR |
hair growth associated |
| chr1_+_151483855 | 0.39 |
ENST00000427934.2 ENST00000271636.7 |
CGN |
cingulin |
| chr11_+_1860200 | 0.37 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr14_+_21525981 | 0.36 |
ENST00000308227.2 |
RNASE8 |
ribonuclease, RNase A family, 8 |
| chr19_+_38755203 | 0.36 |
ENST00000587090.1 ENST00000454580.3 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr1_+_13910479 | 0.35 |
ENST00000509009.1 |
PDPN |
podoplanin |
| chr14_+_21510385 | 0.33 |
ENST00000298690.4 |
RNASE7 |
ribonuclease, RNase A family, 7 |
| chr13_+_78109804 | 0.31 |
ENST00000535157.1 |
SCEL |
sciellin |
| chr13_+_78109884 | 0.31 |
ENST00000377246.3 ENST00000349847.3 |
SCEL |
sciellin |
| chr12_-_52828147 | 0.30 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr17_-_46507537 | 0.29 |
ENST00000336915.6 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr6_-_13487825 | 0.29 |
ENST00000603223.1 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chr1_+_13910757 | 0.29 |
ENST00000376061.4 ENST00000513143.1 |
PDPN |
podoplanin |
| chr1_+_156254070 | 0.29 |
ENST00000405535.2 ENST00000456810.1 |
TMEM79 |
transmembrane protein 79 |
| chr1_+_13910194 | 0.28 |
ENST00000376057.4 ENST00000510906.1 |
PDPN |
podoplanin |
| chr6_-_13487784 | 0.28 |
ENST00000379287.3 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chr3_+_167453493 | 0.27 |
ENST00000295777.5 ENST00000472747.2 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr6_+_121756809 | 0.27 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr14_+_75745477 | 0.27 |
ENST00000303562.4 ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr17_+_34431212 | 0.26 |
ENST00000394495.1 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr8_-_144655141 | 0.25 |
ENST00000398882.3 |
MROH6 |
maestro heat-like repeat family member 6 |
| chr11_+_120081475 | 0.25 |
ENST00000328965.4 |
OAF |
OAF homolog (Drosophila) |
| chr17_+_34430980 | 0.24 |
ENST00000250151.4 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr17_+_47572647 | 0.24 |
ENST00000172229.3 |
NGFR |
nerve growth factor receptor |
| chr14_+_61995722 | 0.24 |
ENST00000556347.1 |
RP11-47I22.4 |
RP11-47I22.4 |
| chr15_+_90319557 | 0.23 |
ENST00000341735.3 |
MESP2 |
mesoderm posterior 2 homolog (mouse) |
| chr15_+_89181974 | 0.23 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr1_-_25256368 | 0.23 |
ENST00000308873.6 |
RUNX3 |
runt-related transcription factor 3 |
| chr3_-_182880541 | 0.22 |
ENST00000470251.1 ENST00000265598.3 |
LAMP3 |
lysosomal-associated membrane protein 3 |
| chr1_-_153521714 | 0.22 |
ENST00000368713.3 |
S100A3 |
S100 calcium binding protein A3 |
| chr2_+_68962014 | 0.22 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr4_+_120133791 | 0.21 |
ENST00000274030.6 |
USP53 |
ubiquitin specific peptidase 53 |
| chr8_+_123793633 | 0.21 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr17_+_30814707 | 0.21 |
ENST00000584792.1 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr5_-_54281491 | 0.21 |
ENST00000381405.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr15_+_89182178 | 0.21 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr12_+_19358228 | 0.21 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chrX_+_135618258 | 0.20 |
ENST00000440515.1 ENST00000456412.1 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr15_+_88120158 | 0.20 |
ENST00000560153.1 |
LINC00052 |
long intergenic non-protein coding RNA 52 |
| chrX_+_56259316 | 0.20 |
ENST00000468660.1 |
KLF8 |
Kruppel-like factor 8 |
| chr8_+_104831472 | 0.19 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr17_-_27229948 | 0.19 |
ENST00000426464.2 |
DHRS13 |
dehydrogenase/reductase (SDR family) member 13 |
| chr5_-_176836577 | 0.18 |
ENST00000253496.3 |
F12 |
coagulation factor XII (Hageman factor) |
| chr1_+_153746683 | 0.18 |
ENST00000271857.2 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr12_-_121342170 | 0.18 |
ENST00000353487.2 |
SPPL3 |
signal peptide peptidase like 3 |
| chr3_+_142315225 | 0.17 |
ENST00000457734.2 ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1 |
plastin 1 |
| chr17_-_27230035 | 0.17 |
ENST00000378895.4 ENST00000394901.3 |
DHRS13 |
dehydrogenase/reductase (SDR family) member 13 |
| chr14_-_91526462 | 0.17 |
ENST00000536315.2 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr5_-_54281407 | 0.16 |
ENST00000381403.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr10_-_131762105 | 0.16 |
ENST00000368648.3 ENST00000355311.5 |
EBF3 |
early B-cell factor 3 |
| chr20_-_22565101 | 0.16 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr17_+_55183261 | 0.16 |
ENST00000576295.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chrX_-_128788914 | 0.16 |
ENST00000429967.1 ENST00000307484.6 |
APLN |
apelin |
| chr7_-_27135591 | 0.16 |
ENST00000343060.4 ENST00000355633.5 |
HOXA1 |
homeobox A1 |
| chr3_+_49726932 | 0.15 |
ENST00000327697.6 ENST00000432042.1 ENST00000454491.1 |
RNF123 |
ring finger protein 123 |
| chr16_-_69385681 | 0.15 |
ENST00000288025.3 |
TMED6 |
transmembrane emp24 protein transport domain containing 6 |
| chr2_+_42275153 | 0.15 |
ENST00000294964.5 |
PKDCC |
protein kinase domain containing, cytoplasmic |
| chr11_-_58345569 | 0.15 |
ENST00000528954.1 ENST00000528489.1 |
LPXN |
leupaxin |
| chr11_-_7847519 | 0.15 |
ENST00000328375.1 |
OR5P3 |
olfactory receptor, family 5, subfamily P, member 3 |
| chr15_+_89182156 | 0.15 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr5_-_134914673 | 0.15 |
ENST00000512158.1 |
CXCL14 |
chemokine (C-X-C motif) ligand 14 |
| chrX_-_130423200 | 0.15 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr17_+_37894179 | 0.14 |
ENST00000577695.1 ENST00000309156.4 ENST00000309185.3 |
GRB7 |
growth factor receptor-bound protein 7 |
| chr7_-_100253993 | 0.14 |
ENST00000461605.1 ENST00000160382.5 |
ACTL6B |
actin-like 6B |
| chr9_-_96717654 | 0.14 |
ENST00000253968.6 |
BARX1 |
BARX homeobox 1 |
| chrX_+_16804544 | 0.14 |
ENST00000380122.5 ENST00000398155.4 |
TXLNG |
taxilin gamma |
| chr1_-_11042094 | 0.13 |
ENST00000377004.4 ENST00000377008.4 |
C1orf127 |
chromosome 1 open reading frame 127 |
| chr12_+_50898881 | 0.13 |
ENST00000301180.5 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr11_+_19138670 | 0.13 |
ENST00000446113.2 ENST00000399351.3 |
ZDHHC13 |
zinc finger, DHHC-type containing 13 |
| chr19_-_41934635 | 0.13 |
ENST00000321702.2 |
B3GNT8 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr1_-_204329013 | 0.13 |
ENST00000272203.3 ENST00000414478.1 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
| chr7_-_138666053 | 0.13 |
ENST00000440172.1 ENST00000422774.1 |
KIAA1549 |
KIAA1549 |
| chr1_-_44482979 | 0.13 |
ENST00000360584.2 ENST00000357730.2 ENST00000528803.1 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr18_-_28742813 | 0.13 |
ENST00000257197.3 ENST00000257198.5 |
DSC1 |
desmocollin 1 |
| chr2_-_56150184 | 0.12 |
ENST00000394554.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr12_-_18890940 | 0.12 |
ENST00000543242.1 ENST00000539072.1 ENST00000541966.1 ENST00000266505.7 ENST00000447925.2 ENST00000435379.1 |
PLCZ1 |
phospholipase C, zeta 1 |
| chr4_-_110651111 | 0.12 |
ENST00000502283.1 |
PLA2G12A |
phospholipase A2, group XIIA |
| chr6_+_30294612 | 0.12 |
ENST00000440271.1 ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39 |
tripartite motif containing 39 |
| chr11_+_35198118 | 0.12 |
ENST00000525211.1 ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr7_-_43965937 | 0.12 |
ENST00000455877.1 ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP |
upregulator of cell proliferation |
| chr11_-_128737163 | 0.12 |
ENST00000324003.3 ENST00000392665.2 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr6_-_30043539 | 0.11 |
ENST00000376751.3 ENST00000244360.6 |
RNF39 |
ring finger protein 39 |
| chr19_-_49622348 | 0.11 |
ENST00000408991.2 |
C19orf73 |
chromosome 19 open reading frame 73 |
| chrX_-_130423386 | 0.11 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr15_+_35270552 | 0.11 |
ENST00000391457.2 |
AC114546.1 |
HCG37415; PRO1914; Uncharacterized protein |
| chr11_-_128737259 | 0.11 |
ENST00000440599.2 ENST00000392666.1 ENST00000324036.3 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr12_+_7055767 | 0.11 |
ENST00000447931.2 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr2_-_129076151 | 0.11 |
ENST00000259241.6 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
| chr7_+_107220660 | 0.11 |
ENST00000465919.1 ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr19_+_10397648 | 0.11 |
ENST00000340992.4 ENST00000393717.2 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chrX_-_49056635 | 0.11 |
ENST00000472598.1 ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP |
synaptophysin |
| chr11_-_34535297 | 0.11 |
ENST00000532417.1 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
| chr11_+_34643600 | 0.11 |
ENST00000530286.1 ENST00000533754.1 |
EHF |
ets homologous factor |
| chr3_+_14860831 | 0.11 |
ENST00000543601.1 |
FGD5 |
FYVE, RhoGEF and PH domain containing 5 |
| chr1_+_165513231 | 0.10 |
ENST00000294818.1 |
LRRC52 |
leucine rich repeat containing 52 |
| chr11_-_67415048 | 0.10 |
ENST00000529256.1 |
ACY3 |
aspartoacylase (aminocyclase) 3 |
| chr7_-_71801980 | 0.10 |
ENST00000329008.5 |
CALN1 |
calneuron 1 |
| chr11_-_123525289 | 0.10 |
ENST00000392770.2 ENST00000299333.3 ENST00000530277.1 |
SCN3B |
sodium channel, voltage-gated, type III, beta subunit |
| chr11_-_59383617 | 0.10 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr4_-_40631859 | 0.10 |
ENST00000295971.7 ENST00000319592.4 |
RBM47 |
RNA binding motif protein 47 |
| chr1_-_207095324 | 0.10 |
ENST00000530505.1 ENST00000367091.3 ENST00000442471.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr6_-_45983549 | 0.10 |
ENST00000544153.1 |
CLIC5 |
chloride intracellular channel 5 |
| chr16_-_11350036 | 0.10 |
ENST00000332029.2 |
SOCS1 |
suppressor of cytokine signaling 1 |
| chr11_+_58390132 | 0.10 |
ENST00000361987.4 |
CNTF |
ciliary neurotrophic factor |
| chr11_-_72385437 | 0.10 |
ENST00000418754.2 ENST00000542969.2 ENST00000334456.5 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr7_-_142120321 | 0.10 |
ENST00000390377.1 |
TRBV7-7 |
T cell receptor beta variable 7-7 |
| chr6_+_30295036 | 0.10 |
ENST00000376659.5 ENST00000428555.1 |
TRIM39 |
tripartite motif containing 39 |
| chr1_+_93544791 | 0.09 |
ENST00000545708.1 ENST00000540243.1 ENST00000370298.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr16_-_2031464 | 0.09 |
ENST00000356120.4 ENST00000354249.4 |
NOXO1 |
NADPH oxidase organizer 1 |
| chr17_+_77020224 | 0.09 |
ENST00000339142.2 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr1_+_93544821 | 0.09 |
ENST00000370303.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr17_+_74372662 | 0.09 |
ENST00000591651.1 ENST00000545180.1 |
SPHK1 |
sphingosine kinase 1 |
| chr17_+_77020325 | 0.09 |
ENST00000311661.4 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr17_-_61996192 | 0.09 |
ENST00000392824.4 |
CSHL1 |
chorionic somatomammotropin hormone-like 1 |
| chr17_-_74533963 | 0.09 |
ENST00000293230.5 |
CYGB |
cytoglobin |
| chr12_+_81101277 | 0.09 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr14_+_24600484 | 0.09 |
ENST00000267426.5 |
FITM1 |
fat storage-inducing transmembrane protein 1 |
| chr17_-_1588101 | 0.09 |
ENST00000577001.1 ENST00000572621.1 ENST00000304992.6 |
PRPF8 |
pre-mRNA processing factor 8 |
| chr19_+_10397621 | 0.09 |
ENST00000380770.3 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr16_-_4465886 | 0.09 |
ENST00000539968.1 |
CORO7 |
coronin 7 |
| chr1_+_27189631 | 0.09 |
ENST00000339276.4 |
SFN |
stratifin |
| chr2_+_96991935 | 0.09 |
ENST00000361124.4 ENST00000420728.1 ENST00000542887.1 |
ITPRIPL1 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
| chr19_-_4302375 | 0.08 |
ENST00000600114.1 ENST00000600349.1 ENST00000595645.1 ENST00000301272.2 |
TMIGD2 |
transmembrane and immunoglobulin domain containing 2 |
| chr11_-_5248294 | 0.08 |
ENST00000335295.4 |
HBB |
hemoglobin, beta |
| chr3_-_49142504 | 0.08 |
ENST00000306125.6 ENST00000420147.2 |
QARS |
glutaminyl-tRNA synthetase |
| chr11_-_34535332 | 0.08 |
ENST00000257832.2 ENST00000429939.2 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
| chr2_+_11696464 | 0.08 |
ENST00000234142.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr20_+_35201993 | 0.08 |
ENST00000373872.4 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chrX_-_11445856 | 0.08 |
ENST00000380736.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr2_+_171673417 | 0.08 |
ENST00000344257.5 |
GAD1 |
glutamate decarboxylase 1 (brain, 67kDa) |
| chr19_+_41949054 | 0.08 |
ENST00000378187.2 |
C19orf69 |
chromosome 19 open reading frame 69 |
| chr20_+_52105495 | 0.08 |
ENST00000439873.2 |
AL354993.1 |
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr10_+_135340859 | 0.08 |
ENST00000252945.3 ENST00000421586.1 ENST00000418356.1 |
CYP2E1 |
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr1_+_15736359 | 0.08 |
ENST00000375980.4 |
EFHD2 |
EF-hand domain family, member D2 |
| chr1_-_205744205 | 0.08 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr3_-_61237050 | 0.07 |
ENST00000476844.1 ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT |
fragile histidine triad |
| chr2_+_171571827 | 0.07 |
ENST00000375281.3 |
SP5 |
Sp5 transcription factor |
| chr17_-_39324424 | 0.07 |
ENST00000391356.2 |
KRTAP4-3 |
keratin associated protein 4-3 |
| chr3_-_52443799 | 0.07 |
ENST00000470173.1 ENST00000296288.5 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr2_+_171673072 | 0.07 |
ENST00000358196.3 ENST00000375272.1 |
GAD1 |
glutamate decarboxylase 1 (brain, 67kDa) |
| chr12_+_7055631 | 0.07 |
ENST00000543115.1 ENST00000399448.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr4_+_81118647 | 0.07 |
ENST00000415738.2 |
PRDM8 |
PR domain containing 8 |
| chr16_-_50715196 | 0.07 |
ENST00000423026.2 |
SNX20 |
sorting nexin 20 |
| chr3_+_134514093 | 0.07 |
ENST00000398015.3 |
EPHB1 |
EPH receptor B1 |
| chr21_+_30502806 | 0.07 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr3_-_49142178 | 0.07 |
ENST00000452739.1 ENST00000414533.1 ENST00000417025.1 |
QARS |
glutaminyl-tRNA synthetase |
| chr17_+_77020146 | 0.07 |
ENST00000579760.1 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr22_+_38597889 | 0.07 |
ENST00000338483.2 ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr6_+_167536230 | 0.07 |
ENST00000341935.5 ENST00000349984.4 |
CCR6 |
chemokine (C-C motif) receptor 6 |
| chr17_-_46703826 | 0.07 |
ENST00000550387.1 ENST00000311177.5 |
HOXB9 |
homeobox B9 |
| chr21_-_46962379 | 0.07 |
ENST00000311124.4 ENST00000380010.4 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
| chr17_+_35849937 | 0.07 |
ENST00000394389.4 |
DUSP14 |
dual specificity phosphatase 14 |
| chr1_+_171750776 | 0.07 |
ENST00000458517.1 ENST00000362019.3 ENST00000367737.5 ENST00000361735.3 |
METTL13 |
methyltransferase like 13 |
| chr3_-_197024394 | 0.07 |
ENST00000434148.1 ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr3_+_14860469 | 0.07 |
ENST00000285046.5 |
FGD5 |
FYVE, RhoGEF and PH domain containing 5 |
| chr2_+_189156638 | 0.07 |
ENST00000410051.1 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr17_-_7297833 | 0.07 |
ENST00000571802.1 ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr8_+_104831554 | 0.07 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chrX_-_130423240 | 0.06 |
ENST00000370910.1 ENST00000370901.4 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr2_+_242127924 | 0.06 |
ENST00000402530.3 ENST00000274979.8 ENST00000402430.3 |
ANO7 |
anoctamin 7 |
| chr3_+_184080790 | 0.06 |
ENST00000430783.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr2_+_191334212 | 0.06 |
ENST00000444317.1 ENST00000535751.1 |
MFSD6 |
major facilitator superfamily domain containing 6 |
| chr4_-_120133661 | 0.06 |
ENST00000503243.1 ENST00000326780.3 |
RP11-455G16.1 |
Uncharacterized protein |
| chr20_-_32891151 | 0.06 |
ENST00000217426.2 |
AHCY |
adenosylhomocysteinase |
| chr4_+_57371509 | 0.06 |
ENST00000360096.2 |
ARL9 |
ADP-ribosylation factor-like 9 |
| chr16_+_66400533 | 0.06 |
ENST00000341529.3 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
| chr9_+_4662282 | 0.06 |
ENST00000381883.2 |
PPAPDC2 |
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr11_+_74660278 | 0.06 |
ENST00000263672.6 ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2 |
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr2_-_56150910 | 0.06 |
ENST00000424836.2 ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr17_-_26898516 | 0.06 |
ENST00000543734.1 ENST00000395346.2 |
PIGS |
phosphatidylinositol glycan anchor biosynthesis, class S |
| chr2_+_29336855 | 0.06 |
ENST00000404424.1 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr1_-_85725316 | 0.06 |
ENST00000344356.5 ENST00000471115.1 |
C1orf52 |
chromosome 1 open reading frame 52 |
| chr19_+_53761545 | 0.06 |
ENST00000341702.3 |
VN1R2 |
vomeronasal 1 receptor 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.2 | 0.9 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.2 | 0.8 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.5 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.8 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.4 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 | 0.6 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.3 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.7 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.4 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.1 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.2 | GO:0021718 | pons maturation(GO:0021586) superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.4 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.2 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.4 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.0 | 0.3 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.0 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.0 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.0 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.1 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.0 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.6 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.2 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.2 | 0.6 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.4 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.5 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 2.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.0 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.0 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |