Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for TCF21
Z-value: 0.59
Transcription factors associated with TCF21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF21
|
ENSG00000118526.6 | TCF21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF21 | hg19_v2_chr6_+_134210243_134210276 | 0.07 | 8.7e-01 | Click! |
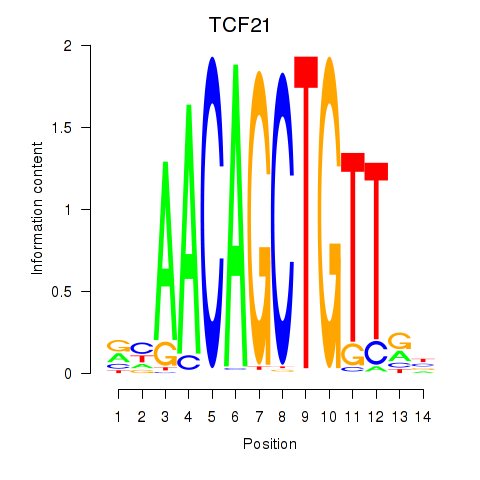
Activity profile of TCF21 motif
Sorted Z-values of TCF21 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF21
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_225811747 | 0.87 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr2_-_158345462 | 0.82 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr5_+_95066823 | 0.59 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr10_-_79398127 | 0.55 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_+_223101757 | 0.43 |
ENST00000284476.6 |
DISP1 |
dispatched homolog 1 (Drosophila) |
| chrX_-_150067069 | 0.42 |
ENST00000466436.1 |
CD99L2 |
CD99 molecule-like 2 |
| chr10_-_79398250 | 0.40 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_-_149889382 | 0.38 |
ENST00000369145.1 ENST00000369146.3 |
SV2A |
synaptic vesicle glycoprotein 2A |
| chr2_+_56411131 | 0.36 |
ENST00000407595.2 |
CCDC85A |
coiled-coil domain containing 85A |
| chrX_-_150067173 | 0.34 |
ENST00000370377.3 ENST00000320893.6 |
CD99L2 |
CD99 molecule-like 2 |
| chr17_+_68165657 | 0.34 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr11_-_59633951 | 0.32 |
ENST00000257264.3 |
TCN1 |
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr1_+_78354297 | 0.31 |
ENST00000334785.7 |
NEXN |
nexilin (F actin binding protein) |
| chr5_-_95158644 | 0.30 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr4_+_114214125 | 0.30 |
ENST00000509550.1 |
ANK2 |
ankyrin 2, neuronal |
| chr9_-_123239632 | 0.29 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr1_-_243326612 | 0.29 |
ENST00000492145.1 ENST00000490813.1 ENST00000464936.1 |
CEP170 |
centrosomal protein 170kDa |
| chr22_+_44319619 | 0.28 |
ENST00000216180.3 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr6_-_31648150 | 0.28 |
ENST00000375858.3 ENST00000383237.4 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr8_+_38585704 | 0.27 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr7_-_92855762 | 0.27 |
ENST00000453812.2 ENST00000394468.2 |
HEPACAM2 |
HEPACAM family member 2 |
| chr1_+_78354175 | 0.27 |
ENST00000401035.3 ENST00000457030.1 ENST00000330010.8 |
NEXN |
nexilin (F actin binding protein) |
| chr17_-_15165854 | 0.26 |
ENST00000395936.1 ENST00000395938.2 |
PMP22 |
peripheral myelin protein 22 |
| chr9_-_104357277 | 0.25 |
ENST00000374806.1 |
PPP3R2 |
protein phosphatase 3, regulatory subunit B, beta |
| chr18_+_3247779 | 0.24 |
ENST00000578611.1 ENST00000583449.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr15_+_93447675 | 0.24 |
ENST00000536619.1 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr1_-_85097431 | 0.24 |
ENST00000327308.3 |
C1orf180 |
chromosome 1 open reading frame 180 |
| chr18_-_55470320 | 0.24 |
ENST00000536015.1 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr10_+_1102721 | 0.23 |
ENST00000263150.4 |
WDR37 |
WD repeat domain 37 |
| chr6_-_31697255 | 0.23 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chrX_+_69509927 | 0.23 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr9_-_89562104 | 0.22 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chr6_+_155470243 | 0.22 |
ENST00000456877.2 ENST00000528391.2 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
| chr9_-_113761720 | 0.22 |
ENST00000541779.1 ENST00000374430.2 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr1_-_31230650 | 0.22 |
ENST00000294507.3 |
LAPTM5 |
lysosomal protein transmembrane 5 |
| chr17_+_72772621 | 0.21 |
ENST00000335464.5 ENST00000417024.2 ENST00000578764.1 ENST00000582773.1 ENST00000582330.1 |
TMEM104 |
transmembrane protein 104 |
| chr2_+_219537015 | 0.21 |
ENST00000440309.1 ENST00000424080.1 |
STK36 |
serine/threonine kinase 36 |
| chr17_+_71228793 | 0.20 |
ENST00000426147.2 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chrX_-_108868390 | 0.20 |
ENST00000372101.2 |
KCNE1L |
KCNE1-like |
| chr2_+_102721023 | 0.20 |
ENST00000409589.1 ENST00000409329.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr17_-_39538550 | 0.19 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chr4_+_154622652 | 0.19 |
ENST00000260010.6 |
TLR2 |
toll-like receptor 2 |
| chr12_-_81992111 | 0.18 |
ENST00000443686.3 ENST00000407050.4 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr3_-_182833863 | 0.18 |
ENST00000492597.1 |
MCCC1 |
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr2_+_219536749 | 0.18 |
ENST00000295709.3 ENST00000392106.2 ENST00000392105.3 ENST00000455724.1 |
STK36 |
serine/threonine kinase 36 |
| chr16_-_31076332 | 0.18 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr13_-_50510622 | 0.18 |
ENST00000378195.2 |
SPRYD7 |
SPRY domain containing 7 |
| chrX_-_106960285 | 0.17 |
ENST00000503515.1 ENST00000372397.2 |
TSC22D3 |
TSC22 domain family, member 3 |
| chrX_+_48432892 | 0.17 |
ENST00000376759.3 ENST00000430348.2 |
RBM3 |
RNA binding motif (RNP1, RRM) protein 3 |
| chr17_+_6347761 | 0.17 |
ENST00000250056.8 ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A |
family with sequence similarity 64, member A |
| chr17_+_6347729 | 0.17 |
ENST00000572447.1 |
FAM64A |
family with sequence similarity 64, member A |
| chr17_-_33864772 | 0.17 |
ENST00000361112.4 |
SLFN12L |
schlafen family member 12-like |
| chr1_+_22963158 | 0.16 |
ENST00000438241.1 |
C1QA |
complement component 1, q subcomponent, A chain |
| chr17_-_7081435 | 0.16 |
ENST00000380920.4 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chrX_+_9431324 | 0.16 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr7_+_20687017 | 0.16 |
ENST00000258738.6 |
ABCB5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_-_85100703 | 0.16 |
ENST00000370624.1 |
C1orf180 |
chromosome 1 open reading frame 180 |
| chr3_-_81792780 | 0.16 |
ENST00000489715.1 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
| chr6_+_26458152 | 0.16 |
ENST00000312541.5 |
BTN2A1 |
butyrophilin, subfamily 2, member A1 |
| chr1_-_155214436 | 0.15 |
ENST00000327247.5 |
GBA |
glucosidase, beta, acid |
| chr1_+_10271674 | 0.15 |
ENST00000377086.1 |
KIF1B |
kinesin family member 1B |
| chr4_+_123844225 | 0.15 |
ENST00000274008.4 |
SPATA5 |
spermatogenesis associated 5 |
| chr10_-_15413035 | 0.15 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chr10_+_94594351 | 0.15 |
ENST00000371552.4 |
EXOC6 |
exocyst complex component 6 |
| chr2_-_224702270 | 0.15 |
ENST00000396654.2 ENST00000396653.2 ENST00000423110.1 ENST00000443700.1 |
AP1S3 |
adaptor-related protein complex 1, sigma 3 subunit |
| chr3_+_141106643 | 0.15 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr9_+_2029019 | 0.14 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_-_31697563 | 0.14 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr22_+_31277661 | 0.14 |
ENST00000454145.1 ENST00000453621.1 ENST00000431368.1 ENST00000535268.1 |
OSBP2 |
oxysterol binding protein 2 |
| chr16_+_77225071 | 0.14 |
ENST00000439557.2 ENST00000545553.1 |
MON1B |
MON1 secretory trafficking family member B |
| chr1_-_150669604 | 0.14 |
ENST00000427665.1 ENST00000540514.1 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr3_+_148583043 | 0.14 |
ENST00000296046.3 |
CPA3 |
carboxypeptidase A3 (mast cell) |
| chr5_+_140723601 | 0.14 |
ENST00000253812.6 |
PCDHGA3 |
protocadherin gamma subfamily A, 3 |
| chr4_+_56262115 | 0.14 |
ENST00000506198.1 ENST00000381334.5 ENST00000542052.1 |
TMEM165 |
transmembrane protein 165 |
| chr22_+_38071615 | 0.14 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr11_-_47270341 | 0.13 |
ENST00000529444.1 ENST00000530453.1 ENST00000537863.1 ENST00000529788.1 ENST00000444355.2 ENST00000527256.1 ENST00000529663.1 ENST00000256997.3 |
ACP2 |
acid phosphatase 2, lysosomal |
| chr12_-_11002063 | 0.13 |
ENST00000544994.1 ENST00000228811.4 ENST00000540107.1 |
PRR4 |
proline rich 4 (lacrimal) |
| chr19_+_50016610 | 0.13 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_-_1643368 | 0.13 |
ENST00000399682.1 |
KRTAP5-4 |
keratin associated protein 5-4 |
| chr3_-_105588231 | 0.13 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr20_+_17680587 | 0.13 |
ENST00000427254.1 ENST00000377805.3 |
BANF2 |
barrier to autointegration factor 2 |
| chr5_-_42811986 | 0.13 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr17_-_67264947 | 0.13 |
ENST00000586811.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr10_-_12084770 | 0.13 |
ENST00000357604.5 |
UPF2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr7_+_66205712 | 0.13 |
ENST00000451741.2 ENST00000442563.1 ENST00000450873.2 ENST00000284957.5 |
KCTD7 RABGEF1 |
potassium channel tetramerization domain containing 7 RAB guanine nucleotide exchange factor (GEF) 1 |
| chr18_+_56807096 | 0.13 |
ENST00000588875.1 |
SEC11C |
SEC11 homolog C (S. cerevisiae) |
| chr15_+_89787180 | 0.13 |
ENST00000300027.8 ENST00000310775.7 ENST00000567891.1 ENST00000564920.1 ENST00000565255.1 ENST00000567996.1 ENST00000451393.2 ENST00000563250.1 |
FANCI |
Fanconi anemia, complementation group I |
| chr20_+_44044717 | 0.13 |
ENST00000279036.6 ENST00000279035.9 ENST00000372689.5 ENST00000545755.1 ENST00000341555.5 ENST00000535404.1 ENST00000543458.2 ENST00000432270.1 |
PIGT |
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr11_+_827553 | 0.13 |
ENST00000528542.2 ENST00000450448.1 |
EFCAB4A |
EF-hand calcium binding domain 4A |
| chr1_-_165414414 | 0.13 |
ENST00000359842.5 |
RXRG |
retinoid X receptor, gamma |
| chr13_-_50510434 | 0.13 |
ENST00000361840.3 |
SPRYD7 |
SPRY domain containing 7 |
| chr6_-_11382478 | 0.12 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr17_+_7341586 | 0.12 |
ENST00000575235.1 |
FGF11 |
fibroblast growth factor 11 |
| chr1_+_12524965 | 0.12 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_-_9129895 | 0.12 |
ENST00000473209.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr2_+_233562015 | 0.11 |
ENST00000427233.1 ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
| chr22_+_45680822 | 0.11 |
ENST00000216211.4 ENST00000396082.2 |
UPK3A |
uroplakin 3A |
| chr11_-_117103208 | 0.11 |
ENST00000320934.3 ENST00000530269.1 |
PCSK7 |
proprotein convertase subtilisin/kexin type 7 |
| chr16_-_46655538 | 0.11 |
ENST00000303383.3 |
SHCBP1 |
SHC SH2-domain binding protein 1 |
| chr13_+_41635617 | 0.11 |
ENST00000542082.1 |
WBP4 |
WW domain binding protein 4 |
| chr19_+_1269324 | 0.11 |
ENST00000589710.1 ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP |
cold inducible RNA binding protein |
| chr7_+_99102573 | 0.11 |
ENST00000394170.2 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr12_-_58329819 | 0.11 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr15_+_65843130 | 0.11 |
ENST00000569894.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chrX_-_103268259 | 0.11 |
ENST00000217926.5 |
H2BFWT |
H2B histone family, member W, testis-specific |
| chr11_+_58910295 | 0.11 |
ENST00000420244.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr20_+_34802295 | 0.11 |
ENST00000432603.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr11_+_58910201 | 0.11 |
ENST00000528737.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr16_-_18441131 | 0.11 |
ENST00000339303.5 |
NPIPA8 |
nuclear pore complex interacting protein family, member A8 |
| chrX_+_78426469 | 0.11 |
ENST00000276077.1 |
GPR174 |
G protein-coupled receptor 174 |
| chr9_+_125137565 | 0.10 |
ENST00000373698.5 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_-_179545059 | 0.10 |
ENST00000367615.4 ENST00000367616.4 |
NPHS2 |
nephrosis 2, idiopathic, steroid-resistant (podocin) |
| chr15_+_63340647 | 0.10 |
ENST00000404484.4 |
TPM1 |
tropomyosin 1 (alpha) |
| chr15_+_63340553 | 0.10 |
ENST00000334895.5 |
TPM1 |
tropomyosin 1 (alpha) |
| chr14_-_53258180 | 0.10 |
ENST00000554230.1 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
| chr6_-_30080876 | 0.10 |
ENST00000376734.3 |
TRIM31 |
tripartite motif containing 31 |
| chr1_+_153600869 | 0.10 |
ENST00000292169.1 ENST00000368696.3 ENST00000436839.1 |
S100A1 |
S100 calcium binding protein A1 |
| chr17_-_53809473 | 0.10 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr2_-_17981462 | 0.10 |
ENST00000402989.1 ENST00000428868.1 |
SMC6 |
structural maintenance of chromosomes 6 |
| chr15_-_79103757 | 0.10 |
ENST00000388820.4 |
ADAMTS7 |
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chr3_+_63263788 | 0.10 |
ENST00000478300.1 |
SYNPR |
synaptoporin |
| chr1_-_147599549 | 0.10 |
ENST00000369228.5 |
NBPF24 |
neuroblastoma breakpoint family, member 24 |
| chr15_+_63340734 | 0.10 |
ENST00000560959.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr19_-_54850417 | 0.10 |
ENST00000291759.4 |
LILRA4 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 |
| chr8_-_92053212 | 0.10 |
ENST00000285419.3 |
TMEM55A |
transmembrane protein 55A |
| chr7_+_99102267 | 0.10 |
ENST00000326775.5 ENST00000451158.1 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr12_-_6960407 | 0.10 |
ENST00000540683.1 ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3 |
cell division cycle associated 3 |
| chr1_-_23857698 | 0.10 |
ENST00000361729.2 |
E2F2 |
E2F transcription factor 2 |
| chr15_+_63340775 | 0.10 |
ENST00000559281.1 ENST00000317516.7 |
TPM1 |
tropomyosin 1 (alpha) |
| chr2_+_220299547 | 0.09 |
ENST00000312358.7 |
SPEG |
SPEG complex locus |
| chr8_+_9413410 | 0.09 |
ENST00000520408.1 ENST00000310430.6 ENST00000522110.1 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr5_+_68462944 | 0.09 |
ENST00000506572.1 |
CCNB1 |
cyclin B1 |
| chr1_+_25757376 | 0.09 |
ENST00000399766.3 ENST00000399763.3 ENST00000374343.4 |
TMEM57 |
transmembrane protein 57 |
| chr7_-_99698338 | 0.09 |
ENST00000354230.3 ENST00000425308.1 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr12_+_8850471 | 0.09 |
ENST00000535829.1 ENST00000357529.3 |
RIMKLB |
ribosomal modification protein rimK-like family member B |
| chr2_-_234763147 | 0.09 |
ENST00000411486.2 ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP |
Holliday junction recognition protein |
| chr20_+_30327063 | 0.09 |
ENST00000300403.6 ENST00000340513.4 |
TPX2 |
TPX2, microtubule-associated |
| chr17_-_4167142 | 0.09 |
ENST00000570535.1 ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chr16_-_31076273 | 0.09 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr15_+_90895471 | 0.09 |
ENST00000354377.3 ENST00000379090.5 |
ZNF774 |
zinc finger protein 774 |
| chrX_+_140982452 | 0.09 |
ENST00000544766.1 |
MAGEC3 |
melanoma antigen family C, 3 |
| chr8_-_71519889 | 0.09 |
ENST00000521425.1 |
TRAM1 |
translocation associated membrane protein 1 |
| chr15_+_63340858 | 0.09 |
ENST00000560615.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr6_+_10528560 | 0.09 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr5_+_68463043 | 0.09 |
ENST00000508407.1 ENST00000505500.1 |
CCNB1 |
cyclin B1 |
| chr17_+_26989109 | 0.09 |
ENST00000314616.6 ENST00000347486.4 |
SUPT6H |
suppressor of Ty 6 homolog (S. cerevisiae) |
| chr11_-_615570 | 0.09 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chr11_+_6947720 | 0.09 |
ENST00000414517.2 |
ZNF215 |
zinc finger protein 215 |
| chr22_-_23922410 | 0.09 |
ENST00000249053.3 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
| chr1_-_226076843 | 0.09 |
ENST00000272134.5 |
LEFTY1 |
left-right determination factor 1 |
| chr1_-_230513367 | 0.09 |
ENST00000321327.2 ENST00000525115.1 |
PGBD5 |
piggyBac transposable element derived 5 |
| chr16_+_16484691 | 0.09 |
ENST00000344087.4 |
NPIPA7 |
nuclear pore complex interacting protein family, member A7 |
| chr20_-_43133491 | 0.08 |
ENST00000411544.1 |
SERINC3 |
serine incorporator 3 |
| chr1_+_144151520 | 0.08 |
ENST00000369372.4 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr14_-_50154921 | 0.08 |
ENST00000553805.2 ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2 |
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr2_-_63815860 | 0.08 |
ENST00000272321.7 ENST00000431065.1 |
WDPCP |
WD repeat containing planar cell polarity effector |
| chr2_-_106054952 | 0.08 |
ENST00000336660.5 ENST00000393352.3 ENST00000607522.1 |
FHL2 |
four and a half LIM domains 2 |
| chr10_+_35415851 | 0.08 |
ENST00000374726.3 |
CREM |
cAMP responsive element modulator |
| chr3_-_10332416 | 0.08 |
ENST00000450603.1 ENST00000449554.2 |
GHRL |
ghrelin/obestatin prepropeptide |
| chr17_-_79805146 | 0.08 |
ENST00000415593.1 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
| chrX_-_52683950 | 0.08 |
ENST00000298181.5 |
SSX7 |
synovial sarcoma, X breakpoint 7 |
| chr2_-_55276320 | 0.08 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chr17_-_39183452 | 0.08 |
ENST00000361883.5 |
KRTAP1-5 |
keratin associated protein 1-5 |
| chr5_-_179050066 | 0.08 |
ENST00000329433.6 ENST00000510411.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr2_-_3595547 | 0.08 |
ENST00000438485.1 |
RP13-512J5.1 |
Uncharacterized protein |
| chr2_-_27593306 | 0.08 |
ENST00000347454.4 |
EIF2B4 |
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr20_-_1165117 | 0.08 |
ENST00000381894.3 |
TMEM74B |
transmembrane protein 74B |
| chr6_-_30658745 | 0.08 |
ENST00000376420.5 ENST00000376421.5 |
NRM |
nurim (nuclear envelope membrane protein) |
| chr1_+_148739443 | 0.08 |
ENST00000417839.1 |
NBPF16 |
neuroblastoma breakpoint family, member 16 |
| chr5_-_36001108 | 0.08 |
ENST00000333811.4 |
UGT3A1 |
UDP glycosyltransferase 3 family, polypeptide A1 |
| chr6_-_116833500 | 0.08 |
ENST00000356128.4 |
TRAPPC3L |
trafficking protein particle complex 3-like |
| chr9_-_130541017 | 0.07 |
ENST00000314830.8 |
SH2D3C |
SH2 domain containing 3C |
| chr5_+_170288856 | 0.07 |
ENST00000523189.1 |
RANBP17 |
RAN binding protein 17 |
| chr6_-_128841503 | 0.07 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr5_-_140700322 | 0.07 |
ENST00000313368.5 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr11_+_32112431 | 0.07 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr4_+_106816644 | 0.07 |
ENST00000506666.1 ENST00000503451.1 |
NPNT |
nephronectin |
| chrM_+_7586 | 0.07 |
ENST00000361739.1 |
MT-CO2 |
mitochondrially encoded cytochrome c oxidase II |
| chr5_+_68462837 | 0.07 |
ENST00000256442.5 |
CCNB1 |
cyclin B1 |
| chr3_+_122296443 | 0.07 |
ENST00000464300.2 |
PARP15 |
poly (ADP-ribose) polymerase family, member 15 |
| chr1_+_22328144 | 0.07 |
ENST00000290122.3 ENST00000374663.1 |
CELA3A |
chymotrypsin-like elastase family, member 3A |
| chr20_-_3662866 | 0.07 |
ENST00000356518.2 ENST00000379861.4 |
ADAM33 |
ADAM metallopeptidase domain 33 |
| chr18_+_158513 | 0.07 |
ENST00000400266.3 ENST00000580410.1 ENST00000383589.2 ENST00000261601.7 |
USP14 |
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr10_+_35415719 | 0.07 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr2_-_122494487 | 0.07 |
ENST00000451734.1 ENST00000285814.4 |
MKI67IP |
nucleolar protein interacting with the FHA domain of MKI67 |
| chr7_+_142498725 | 0.07 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr12_+_122326662 | 0.07 |
ENST00000261817.2 ENST00000538613.1 ENST00000542602.1 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr1_+_79115503 | 0.07 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chrX_-_16887963 | 0.07 |
ENST00000380084.4 |
RBBP7 |
retinoblastoma binding protein 7 |
| chr22_+_40742512 | 0.07 |
ENST00000454266.2 ENST00000342312.6 |
ADSL |
adenylosuccinate lyase |
| chr5_+_49962772 | 0.07 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr11_+_125496619 | 0.07 |
ENST00000532669.1 ENST00000278916.3 |
CHEK1 |
checkpoint kinase 1 |
| chr7_-_99569468 | 0.07 |
ENST00000419575.1 |
AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
| chr8_+_66955648 | 0.07 |
ENST00000522619.1 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr5_-_98262240 | 0.07 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chr19_-_49828438 | 0.07 |
ENST00000454748.3 ENST00000598828.1 ENST00000335875.4 |
SLC6A16 |
solute carrier family 6, member 16 |
| chr12_-_52585765 | 0.07 |
ENST00000313234.5 ENST00000394815.2 |
KRT80 |
keratin 80 |
| chr19_-_50400212 | 0.07 |
ENST00000391826.2 |
IL4I1 |
interleukin 4 induced 1 |
| chr17_+_77681075 | 0.07 |
ENST00000397549.2 |
CTD-2116F7.1 |
CTD-2116F7.1 |
| chr2_+_231280954 | 0.07 |
ENST00000409824.1 ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100 |
SP100 nuclear antigen |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.0 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.3 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.1 | 0.4 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.9 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.2 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.2 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:1990768 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0070213 | negative regulation of sister chromatid cohesion(GO:0045875) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.2 | GO:1990539 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0072312 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.0 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.0 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0090521 | regulation of embryonic cell shape(GO:0016476) septin cytoskeleton organization(GO:0032185) glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.0 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.2 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.3 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |