Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
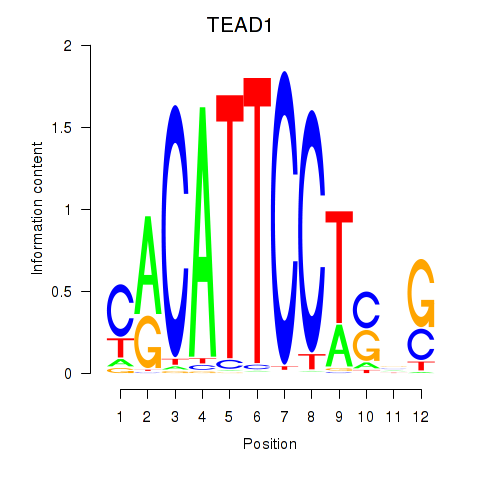
Results for TEAD3_TEAD1
Z-value: 0.38
Transcription factors associated with TEAD3_TEAD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD3
|
ENSG00000007866.14 | TEAD3 |
|
TEAD1
|
ENSG00000187079.10 | TEAD1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD3 | hg19_v2_chr6_-_35464727_35464738, hg19_v2_chr6_-_35464817_35464894 | 0.15 | 7.2e-01 | Click! |
| TEAD1 | hg19_v2_chr11_+_12766583_12766592, hg19_v2_chr11_+_12696102_12696164 | 0.14 | 7.4e-01 | Click! |
Activity profile of TEAD3_TEAD1 motif
Sorted Z-values of TEAD3_TEAD1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD3_TEAD1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_132272504 | 1.70 |
ENST00000367976.3 |
CTGF |
connective tissue growth factor |
| chr5_-_39425068 | 1.48 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr19_+_45409011 | 1.30 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr19_+_34972543 | 1.28 |
ENST00000590071.2 |
WTIP |
Wilms tumor 1 interacting protein |
| chr8_-_108510224 | 1.28 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr1_-_1293904 | 1.13 |
ENST00000309212.6 ENST00000342753.4 ENST00000445648.2 |
MXRA8 |
matrix-remodelling associated 8 |
| chr11_-_111781454 | 1.12 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr16_+_29823427 | 1.00 |
ENST00000358758.7 ENST00000567659.1 ENST00000572820.1 |
PRRT2 |
proline-rich transmembrane protein 2 |
| chr3_-_112360116 | 0.93 |
ENST00000206423.3 ENST00000439685.2 |
CCDC80 |
coiled-coil domain containing 80 |
| chr9_-_139891165 | 0.87 |
ENST00000494426.1 |
CLIC3 |
chloride intracellular channel 3 |
| chr16_+_29823552 | 0.75 |
ENST00000300797.6 |
PRRT2 |
proline-rich transmembrane protein 2 |
| chr12_-_96184533 | 0.73 |
ENST00000343702.4 ENST00000344911.4 |
NTN4 |
netrin 4 |
| chr16_+_30907927 | 0.69 |
ENST00000279804.2 ENST00000395019.3 |
CTF1 |
cardiotrophin 1 |
| chr9_+_118916082 | 0.68 |
ENST00000328252.3 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr5_-_55290773 | 0.66 |
ENST00000502326.3 ENST00000381298.2 |
IL6ST |
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr5_-_39425290 | 0.66 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_-_186733363 | 0.66 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr3_-_114343039 | 0.64 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_-_39425222 | 0.64 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr8_+_70378852 | 0.64 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr11_-_111781554 | 0.62 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr4_-_175443943 | 0.61 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr15_+_33010175 | 0.61 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr12_-_7245125 | 0.60 |
ENST00000542285.1 ENST00000540610.1 |
C1R |
complement component 1, r subcomponent |
| chr11_-_111781610 | 0.57 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr11_-_111782696 | 0.57 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr12_-_47219733 | 0.57 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chr10_-_90712520 | 0.56 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr16_+_30387141 | 0.55 |
ENST00000566955.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr8_-_120605194 | 0.55 |
ENST00000522167.1 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr11_-_111782484 | 0.54 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr20_+_35169885 | 0.54 |
ENST00000279022.2 ENST00000346786.2 |
MYL9 |
myosin, light chain 9, regulatory |
| chr18_-_3247084 | 0.53 |
ENST00000609924.1 |
RP13-270P17.3 |
RP13-270P17.3 |
| chr2_+_210444748 | 0.53 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr20_+_33146510 | 0.52 |
ENST00000397709.1 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
| chr14_+_76044934 | 0.51 |
ENST00000238667.4 |
FLVCR2 |
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr1_+_78354297 | 0.51 |
ENST00000334785.7 |
NEXN |
nexilin (F actin binding protein) |
| chr3_-_191000172 | 0.51 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr15_-_48937982 | 0.48 |
ENST00000316623.5 |
FBN1 |
fibrillin 1 |
| chr13_+_32605437 | 0.47 |
ENST00000380250.3 |
FRY |
furry homolog (Drosophila) |
| chr15_+_62853562 | 0.46 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr16_-_74455290 | 0.45 |
ENST00000339953.5 |
CLEC18B |
C-type lectin domain family 18, member B |
| chr5_+_131630117 | 0.45 |
ENST00000200652.3 |
SLC22A4 |
solute carrier family 22 (organic cation/zwitterion transporter), member 4 |
| chr11_+_46316677 | 0.45 |
ENST00000534787.1 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr1_-_3447967 | 0.44 |
ENST00000294599.4 |
MEGF6 |
multiple EGF-like-domains 6 |
| chr11_+_58390132 | 0.44 |
ENST00000361987.4 |
CNTF |
ciliary neurotrophic factor |
| chr19_-_44388116 | 0.44 |
ENST00000587539.1 |
ZNF404 |
zinc finger protein 404 |
| chr1_+_114522049 | 0.44 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr17_-_39093672 | 0.43 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chr15_+_41549105 | 0.42 |
ENST00000560965.1 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr11_+_46299199 | 0.40 |
ENST00000529193.1 ENST00000288400.3 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr19_-_51472031 | 0.40 |
ENST00000391808.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr1_+_78354175 | 0.40 |
ENST00000401035.3 ENST00000457030.1 ENST00000330010.8 |
NEXN |
nexilin (F actin binding protein) |
| chr1_+_150229554 | 0.39 |
ENST00000369111.4 |
CA14 |
carbonic anhydrase XIV |
| chr16_-_122619 | 0.39 |
ENST00000262316.6 |
RHBDF1 |
rhomboid 5 homolog 1 (Drosophila) |
| chr19_-_51472222 | 0.39 |
ENST00000376851.3 |
KLK6 |
kallikrein-related peptidase 6 |
| chr4_-_186877502 | 0.39 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr11_+_130542851 | 0.39 |
ENST00000317019.2 |
C11orf44 |
chromosome 11 open reading frame 44 |
| chr14_-_60337684 | 0.37 |
ENST00000267484.5 |
RTN1 |
reticulon 1 |
| chr8_+_97597148 | 0.37 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr2_-_190044480 | 0.35 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr11_-_47470703 | 0.35 |
ENST00000298854.2 |
RAPSN |
receptor-associated protein of the synapse |
| chr3_-_119379719 | 0.35 |
ENST00000493094.1 |
POPDC2 |
popeye domain containing 2 |
| chr6_-_128841503 | 0.35 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr9_-_20622478 | 0.35 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr11_-_47470591 | 0.34 |
ENST00000524487.1 |
RAPSN |
receptor-associated protein of the synapse |
| chr9_-_73483958 | 0.33 |
ENST00000377101.1 ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr15_+_63334831 | 0.32 |
ENST00000288398.6 ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr3_-_119379427 | 0.32 |
ENST00000264231.3 ENST00000468801.1 ENST00000538678.1 |
POPDC2 |
popeye domain containing 2 |
| chr8_-_38325219 | 0.32 |
ENST00000533668.1 ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr17_-_27949911 | 0.31 |
ENST00000492276.2 ENST00000345068.5 ENST00000584602.1 |
CORO6 |
coronin 6 |
| chr19_-_14048804 | 0.31 |
ENST00000254320.3 ENST00000586075.1 |
PODNL1 |
podocan-like 1 |
| chr11_+_48002076 | 0.30 |
ENST00000418331.2 ENST00000440289.2 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
| chr2_-_111230393 | 0.29 |
ENST00000447537.2 ENST00000413601.2 |
LIMS3L LIMS3L |
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein LIM and senescent cell antigen-like domains 3-like |
| chr11_-_47470682 | 0.29 |
ENST00000529341.1 ENST00000352508.3 |
RAPSN |
receptor-associated protein of the synapse |
| chr10_-_17659234 | 0.28 |
ENST00000466335.1 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr12_-_63328817 | 0.28 |
ENST00000228705.6 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr6_+_108881012 | 0.28 |
ENST00000343882.6 |
FOXO3 |
forkhead box O3 |
| chr4_-_186877806 | 0.28 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr7_-_150946015 | 0.27 |
ENST00000262188.8 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr13_-_67804445 | 0.27 |
ENST00000456367.1 ENST00000377861.3 ENST00000544246.1 |
PCDH9 |
protocadherin 9 |
| chr1_+_196621156 | 0.27 |
ENST00000359637.2 |
CFH |
complement factor H |
| chr1_-_201915590 | 0.26 |
ENST00000367288.4 |
LMOD1 |
leiomodin 1 (smooth muscle) |
| chr13_-_33760216 | 0.26 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr2_+_110656005 | 0.26 |
ENST00000437679.2 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
| chr1_+_196621002 | 0.26 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr3_-_9994021 | 0.26 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chr11_+_65190245 | 0.26 |
ENST00000499732.1 ENST00000501122.2 ENST00000601801.1 |
NEAT1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr2_-_175499294 | 0.25 |
ENST00000392547.2 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr8_+_38831683 | 0.25 |
ENST00000302495.4 |
HTRA4 |
HtrA serine peptidase 4 |
| chr7_-_137686791 | 0.25 |
ENST00000452463.1 ENST00000330387.6 ENST00000456390.1 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
| chr6_-_111804905 | 0.25 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr22_+_24577183 | 0.24 |
ENST00000358321.3 |
SUSD2 |
sushi domain containing 2 |
| chr11_-_62521614 | 0.24 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chr17_+_20059358 | 0.24 |
ENST00000536879.1 ENST00000395522.2 ENST00000395525.3 |
SPECC1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr9_+_18474098 | 0.24 |
ENST00000327883.7 ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1 |
ADAMTS-like 1 |
| chr14_-_89021077 | 0.23 |
ENST00000556564.1 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
| chr4_-_186732048 | 0.23 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chrX_+_99899180 | 0.23 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr6_-_152489484 | 0.23 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr18_-_25616519 | 0.23 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr16_-_57570450 | 0.23 |
ENST00000258214.2 |
CCDC102A |
coiled-coil domain containing 102A |
| chr12_-_7281469 | 0.23 |
ENST00000542370.1 ENST00000266560.3 |
RBP5 |
retinol binding protein 5, cellular |
| chr16_+_30386098 | 0.23 |
ENST00000322861.7 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr9_+_72658490 | 0.23 |
ENST00000377182.4 |
MAMDC2 |
MAM domain containing 2 |
| chr18_+_39535152 | 0.22 |
ENST00000262039.4 ENST00000398870.3 ENST00000586545.1 |
PIK3C3 |
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr10_-_106240032 | 0.22 |
ENST00000447860.1 |
RP11-127O4.3 |
RP11-127O4.3 |
| chr2_+_102759199 | 0.22 |
ENST00000409288.1 ENST00000410023.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr17_+_73717407 | 0.22 |
ENST00000579662.1 |
ITGB4 |
integrin, beta 4 |
| chrX_-_83757399 | 0.22 |
ENST00000373177.2 ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX |
highly divergent homeobox |
| chr12_-_122241812 | 0.21 |
ENST00000538335.1 |
AC084018.1 |
AC084018.1 |
| chr11_-_1785139 | 0.21 |
ENST00000236671.2 |
CTSD |
cathepsin D |
| chr4_+_166300084 | 0.21 |
ENST00000402744.4 |
CPE |
carboxypeptidase E |
| chr7_+_134430212 | 0.21 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr13_+_76210448 | 0.21 |
ENST00000377499.5 |
LMO7 |
LIM domain 7 |
| chr1_+_74701062 | 0.21 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr17_+_73717516 | 0.21 |
ENST00000200181.3 ENST00000339591.3 |
ITGB4 |
integrin, beta 4 |
| chr19_-_14049184 | 0.21 |
ENST00000339560.5 |
PODNL1 |
podocan-like 1 |
| chr3_+_9944492 | 0.20 |
ENST00000383814.3 ENST00000454190.2 ENST00000454992.1 |
IL17RE |
interleukin 17 receptor E |
| chr6_+_123038689 | 0.20 |
ENST00000354275.2 ENST00000368446.1 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr17_+_73717551 | 0.19 |
ENST00000450894.3 |
ITGB4 |
integrin, beta 4 |
| chr13_-_86373536 | 0.19 |
ENST00000400286.2 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
| chrX_-_142722897 | 0.19 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr19_-_6375860 | 0.19 |
ENST00000245810.1 |
PSPN |
persephin |
| chr17_-_21156578 | 0.19 |
ENST00000399011.2 ENST00000468196.1 |
C17orf103 |
chromosome 17 open reading frame 103 |
| chr2_+_228029281 | 0.19 |
ENST00000396578.3 |
COL4A3 |
collagen, type IV, alpha 3 (Goodpasture antigen) |
| chr12_+_80750134 | 0.19 |
ENST00000546620.1 |
OTOGL |
otogelin-like |
| chr12_-_104443890 | 0.19 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chr3_+_38347427 | 0.19 |
ENST00000273173.4 |
SLC22A14 |
solute carrier family 22, member 14 |
| chr16_-_72206034 | 0.19 |
ENST00000537465.1 ENST00000237353.10 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
| chr11_-_62911693 | 0.19 |
ENST00000417740.1 ENST00000326192.5 |
SLC22A24 |
solute carrier family 22, member 24 |
| chr16_-_31076332 | 0.18 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr8_-_91657740 | 0.18 |
ENST00000422900.1 |
TMEM64 |
transmembrane protein 64 |
| chr2_+_228678550 | 0.18 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr20_+_44044717 | 0.18 |
ENST00000279036.6 ENST00000279035.9 ENST00000372689.5 ENST00000545755.1 ENST00000341555.5 ENST00000535404.1 ENST00000543458.2 ENST00000432270.1 |
PIGT |
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr4_+_41614720 | 0.18 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr10_+_99344071 | 0.18 |
ENST00000370647.4 ENST00000370646.4 |
HOGA1 |
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr5_-_95158644 | 0.18 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr17_-_79105734 | 0.18 |
ENST00000417379.1 |
AATK |
apoptosis-associated tyrosine kinase |
| chr1_+_178310581 | 0.17 |
ENST00000462775.1 |
RASAL2 |
RAS protein activator like 2 |
| chr16_+_69985083 | 0.17 |
ENST00000288040.6 ENST00000449317.2 |
CLEC18A |
C-type lectin domain family 18, member A |
| chr5_+_140868717 | 0.17 |
ENST00000252087.1 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
| chr2_+_145780739 | 0.17 |
ENST00000597173.1 ENST00000602108.1 ENST00000420472.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr2_-_26864228 | 0.17 |
ENST00000288861.4 |
CIB4 |
calcium and integrin binding family member 4 |
| chr5_+_42423872 | 0.17 |
ENST00000230882.4 ENST00000357703.3 |
GHR |
growth hormone receptor |
| chr4_-_119274121 | 0.17 |
ENST00000296498.3 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
| chr9_-_73736511 | 0.17 |
ENST00000377110.3 ENST00000377111.2 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chrX_+_48455866 | 0.17 |
ENST00000376729.5 ENST00000218056.5 |
WDR13 |
WD repeat domain 13 |
| chr15_-_37391614 | 0.17 |
ENST00000219869.9 |
MEIS2 |
Meis homeobox 2 |
| chr3_+_141043050 | 0.17 |
ENST00000509842.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr19_-_4043154 | 0.17 |
ENST00000535853.1 |
AC016586.1 |
AC016586.1 |
| chr3_+_9944303 | 0.17 |
ENST00000421412.1 ENST00000295980.3 |
IL17RE |
interleukin 17 receptor E |
| chr6_-_30899924 | 0.17 |
ENST00000359086.3 |
SFTA2 |
surfactant associated 2 |
| chr16_+_70207686 | 0.16 |
ENST00000541793.2 ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C |
C-type lectin domain family 18, member C |
| chr2_-_183387430 | 0.16 |
ENST00000410103.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr9_+_131684562 | 0.16 |
ENST00000421063.2 |
PHYHD1 |
phytanoyl-CoA dioxygenase domain containing 1 |
| chr8_+_38585704 | 0.16 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr19_+_45349630 | 0.16 |
ENST00000252483.5 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr10_+_81891416 | 0.16 |
ENST00000372270.2 |
PLAC9 |
placenta-specific 9 |
| chr6_+_30850697 | 0.16 |
ENST00000509639.1 ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr4_-_70725856 | 0.16 |
ENST00000226444.3 |
SULT1E1 |
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr11_+_22689648 | 0.16 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chr5_-_146781153 | 0.16 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr7_-_100881041 | 0.16 |
ENST00000412417.1 ENST00000414035.1 |
CLDN15 |
claudin 15 |
| chr5_+_49962772 | 0.16 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr3_-_62358690 | 0.16 |
ENST00000475839.1 |
FEZF2 |
FEZ family zinc finger 2 |
| chr5_-_95158375 | 0.16 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chr17_-_7145106 | 0.16 |
ENST00000577035.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr15_+_43809797 | 0.16 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr15_-_44069741 | 0.15 |
ENST00000319359.3 |
ELL3 |
elongation factor RNA polymerase II-like 3 |
| chr5_+_112074029 | 0.15 |
ENST00000512211.2 |
APC |
adenomatous polyposis coli |
| chr7_+_134464376 | 0.15 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr16_-_31076273 | 0.15 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr5_-_177423243 | 0.15 |
ENST00000308304.2 |
PROP1 |
PROP paired-like homeobox 1 |
| chr19_+_15218180 | 0.15 |
ENST00000342784.2 ENST00000597977.1 ENST00000600440.1 |
SYDE1 |
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr5_+_43121698 | 0.15 |
ENST00000505606.2 ENST00000509634.1 ENST00000509341.1 |
ZNF131 |
zinc finger protein 131 |
| chr12_-_14967095 | 0.15 |
ENST00000316048.2 |
SMCO3 |
single-pass membrane protein with coiled-coil domains 3 |
| chr16_+_1578674 | 0.15 |
ENST00000253934.5 |
TMEM204 |
transmembrane protein 204 |
| chr19_+_39903185 | 0.15 |
ENST00000409794.3 |
PLEKHG2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr7_+_128470431 | 0.15 |
ENST00000325888.8 ENST00000346177.6 |
FLNC |
filamin C, gamma |
| chr3_-_155394152 | 0.14 |
ENST00000494598.1 |
PLCH1 |
phospholipase C, eta 1 |
| chr11_-_124190184 | 0.14 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chr14_-_77495007 | 0.14 |
ENST00000238647.3 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
| chr10_-_92681033 | 0.14 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr13_+_46276441 | 0.14 |
ENST00000310521.1 ENST00000533564.1 |
SPERT |
spermatid associated |
| chr21_+_17566643 | 0.14 |
ENST00000419952.1 ENST00000445461.2 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr5_+_54455946 | 0.14 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr12_-_45307711 | 0.14 |
ENST00000333837.4 ENST00000551949.1 |
NELL2 |
NEL-like 2 (chicken) |
| chr16_+_69984810 | 0.14 |
ENST00000393701.2 ENST00000568461.1 |
CLEC18A |
C-type lectin domain family 18, member A |
| chr9_-_113341985 | 0.14 |
ENST00000374469.1 |
SVEP1 |
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr5_+_118691706 | 0.14 |
ENST00000415806.2 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
| chr1_-_205290865 | 0.14 |
ENST00000367157.3 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
| chr1_-_76398077 | 0.13 |
ENST00000284142.6 |
ASB17 |
ankyrin repeat and SOCS box containing 17 |
| chr7_+_134464414 | 0.13 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr5_-_180688105 | 0.13 |
ENST00000327767.4 |
TRIM52 |
tripartite motif containing 52 |
| chr8_-_57359131 | 0.13 |
ENST00000518974.1 ENST00000523051.1 ENST00000518770.1 ENST00000451791.2 |
PENK |
proenkephalin |
| chr1_-_154474589 | 0.13 |
ENST00000304760.2 |
SHE |
Src homology 2 domain containing E |
| chr18_-_3219847 | 0.13 |
ENST00000261606.7 |
MYOM1 |
myomesin 1 |
| chr12_-_10151773 | 0.13 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr9_+_139873264 | 0.13 |
ENST00000446677.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.6 | 1.7 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.4 | 1.3 | GO:2000646 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 1.0 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.3 | 1.5 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 0.6 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 3.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.3 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 0.8 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.5 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.5 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 | 1.6 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 0.2 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.6 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 1.1 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.6 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.1 | 0.3 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.2 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.4 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.5 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:2001202 | negative regulation of transforming growth factor-beta secretion(GO:2001202) |
| 0.0 | 1.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.6 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0060435 | branchiomeric skeletal muscle development(GO:0014707) bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:0072229 | nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.4 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.2 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0060516 | primary prostatic bud elongation(GO:0060516) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0072615 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.0 | GO:0003011 | diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0039506 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.5 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.4 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0043605 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.0 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.0 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0071694 | sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.0 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.0 | 0.0 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.1 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 2.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 3.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.0 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 1.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0036025 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.4 | GO:0032059 | muscle thin filament tropomyosin(GO:0005862) bleb(GO:0032059) |
| 0.0 | 1.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.3 | 0.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 0.6 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 2.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.4 | GO:0008513 | secondary active organic cation transmembrane transporter activity(GO:0008513) |
| 0.1 | 3.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 2.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 1.0 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.3 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 2.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 1.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.0 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |