Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for TEAD4
Z-value: 0.41
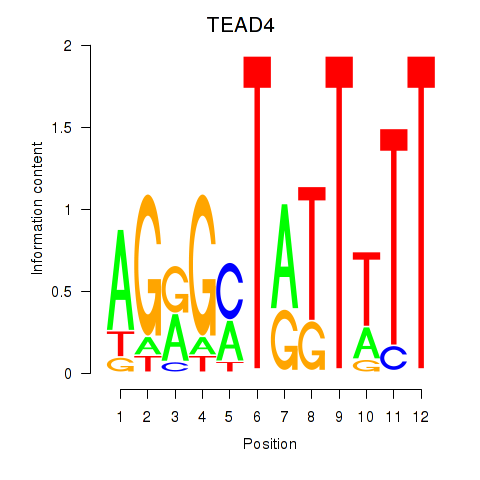
Transcription factors associated with TEAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD4
|
ENSG00000197905.4 | TEAD4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD4 | hg19_v2_chr12_+_3069037_3069119, hg19_v2_chr12_+_3068466_3068496, hg19_v2_chr12_+_3068544_3068597 | -0.36 | 3.7e-01 | Click! |
Activity profile of TEAD4 motif
Sorted Z-values of TEAD4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_161056802 | 0.66 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr2_-_161056762 | 0.60 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr1_+_153004800 | 0.57 |
ENST00000392661.3 |
SPRR1B |
small proline-rich protein 1B |
| chr3_+_189349162 | 0.39 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr7_+_147830776 | 0.36 |
ENST00000538075.1 |
CNTNAP2 |
contactin associated protein-like 2 |
| chr2_-_235405679 | 0.33 |
ENST00000390645.2 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr18_+_61254570 | 0.32 |
ENST00000344731.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr4_-_110723134 | 0.27 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr4_-_89152474 | 0.27 |
ENST00000515655.1 |
ABCG2 |
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr11_-_118134997 | 0.27 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr6_+_125540951 | 0.26 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr1_+_156252708 | 0.25 |
ENST00000295694.5 ENST00000357501.2 |
TMEM79 |
transmembrane protein 79 |
| chr5_+_140261703 | 0.25 |
ENST00000409494.1 ENST00000289272.2 |
PCDHA13 |
protocadherin alpha 13 |
| chr19_-_51466681 | 0.24 |
ENST00000456750.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr12_+_41831485 | 0.24 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr9_-_104249400 | 0.23 |
ENST00000374848.3 |
TMEM246 |
transmembrane protein 246 |
| chr3_+_111805182 | 0.22 |
ENST00000430855.1 ENST00000431717.2 ENST00000264848.5 |
C3orf52 |
chromosome 3 open reading frame 52 |
| chr4_-_110723335 | 0.22 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr1_-_175162048 | 0.22 |
ENST00000444639.1 |
KIAA0040 |
KIAA0040 |
| chr1_+_43735646 | 0.22 |
ENST00000439858.1 |
TMEM125 |
transmembrane protein 125 |
| chr1_+_43735678 | 0.22 |
ENST00000432792.2 |
TMEM125 |
transmembrane protein 125 |
| chr3_+_98699880 | 0.21 |
ENST00000473756.1 |
LINC00973 |
long intergenic non-protein coding RNA 973 |
| chr4_-_110723194 | 0.21 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr7_-_22234381 | 0.20 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr11_-_51412448 | 0.19 |
ENST00000319760.6 |
OR4A5 |
olfactory receptor, family 4, subfamily A, member 5 |
| chr7_-_41742697 | 0.18 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr11_+_10476851 | 0.18 |
ENST00000396553.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr11_-_79151695 | 0.18 |
ENST00000278550.7 |
TENM4 |
teneurin transmembrane protein 4 |
| chrX_-_33146477 | 0.16 |
ENST00000378677.2 |
DMD |
dystrophin |
| chr7_-_16872932 | 0.16 |
ENST00000419572.2 ENST00000412973.1 |
AGR2 |
anterior gradient 2 |
| chr7_+_143701022 | 0.13 |
ENST00000408922.2 |
OR6B1 |
olfactory receptor, family 6, subfamily B, member 1 |
| chr10_-_61513146 | 0.12 |
ENST00000430431.1 |
LINC00948 |
long intergenic non-protein coding RNA 948 |
| chr2_+_1418154 | 0.11 |
ENST00000423320.1 ENST00000382198.1 |
TPO |
thyroid peroxidase |
| chr18_-_67873078 | 0.11 |
ENST00000255674.6 |
RTTN |
rotatin |
| chr4_+_41614720 | 0.11 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr4_-_143227088 | 0.11 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_161676983 | 0.11 |
ENST00000367957.2 |
FCRLA |
Fc receptor-like A |
| chr1_+_161676739 | 0.11 |
ENST00000236938.6 ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA |
Fc receptor-like A |
| chr4_-_143226979 | 0.10 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_107683436 | 0.10 |
ENST00000370068.1 |
NTNG1 |
netrin G1 |
| chr12_-_131200810 | 0.10 |
ENST00000536002.1 ENST00000544034.1 |
RIMBP2 RP11-662M24.2 |
RIMS binding protein 2 RP11-662M24.2 |
| chr10_-_50970322 | 0.10 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr1_-_60392452 | 0.10 |
ENST00000371204.3 |
CYP2J2 |
cytochrome P450, family 2, subfamily J, polypeptide 2 |
| chr7_+_150811705 | 0.10 |
ENST00000335367.3 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr10_-_50970382 | 0.10 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr19_+_41698927 | 0.09 |
ENST00000310054.4 |
CYP2S1 |
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr18_-_67872891 | 0.09 |
ENST00000454359.1 ENST00000437017.1 |
RTTN |
rotatin |
| chrX_+_21392873 | 0.09 |
ENST00000379510.3 |
CNKSR2 |
connector enhancer of kinase suppressor of Ras 2 |
| chr5_-_54281491 | 0.09 |
ENST00000381405.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr11_+_62009653 | 0.09 |
ENST00000244926.3 |
SCGB1D2 |
secretoglobin, family 1D, member 2 |
| chr3_+_123813543 | 0.09 |
ENST00000360013.3 |
KALRN |
kalirin, RhoGEF kinase |
| chrX_+_79675965 | 0.09 |
ENST00000308293.5 |
FAM46D |
family with sequence similarity 46, member D |
| chrX_+_49832231 | 0.09 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr10_-_61513201 | 0.08 |
ENST00000414264.1 ENST00000594536.1 |
LINC00948 |
long intergenic non-protein coding RNA 948 |
| chr7_-_82073109 | 0.08 |
ENST00000356860.3 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr12_+_48876275 | 0.08 |
ENST00000314014.2 |
C12orf54 |
chromosome 12 open reading frame 54 |
| chr7_-_82073031 | 0.07 |
ENST00000356253.5 ENST00000423588.1 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr2_+_182850743 | 0.07 |
ENST00000409702.1 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr7_-_37024665 | 0.07 |
ENST00000396040.2 |
ELMO1 |
engulfment and cell motility 1 |
| chr12_+_20963647 | 0.06 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr10_-_62149433 | 0.06 |
ENST00000280772.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr7_-_107880508 | 0.06 |
ENST00000425651.2 |
NRCAM |
neuronal cell adhesion molecule |
| chr12_+_20963632 | 0.06 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr19_+_9361606 | 0.06 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chr2_+_238768187 | 0.06 |
ENST00000254661.4 ENST00000409726.1 |
RAMP1 |
receptor (G protein-coupled) activity modifying protein 1 |
| chr10_-_75410771 | 0.06 |
ENST00000372873.4 |
SYNPO2L |
synaptopodin 2-like |
| chr1_+_236557569 | 0.06 |
ENST00000334232.4 |
EDARADD |
EDAR-associated death domain |
| chr1_+_66458072 | 0.06 |
ENST00000423207.2 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr4_+_96012614 | 0.06 |
ENST00000264568.4 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chrX_-_77914825 | 0.06 |
ENST00000321110.1 |
ZCCHC5 |
zinc finger, CCHC domain containing 5 |
| chr7_-_116963334 | 0.06 |
ENST00000265441.3 |
WNT2 |
wingless-type MMTV integration site family member 2 |
| chr10_-_13276329 | 0.05 |
ENST00000378681.3 ENST00000463405.2 |
UCMA |
upper zone of growth plate and cartilage matrix associated |
| chr2_-_231989808 | 0.05 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr13_-_99910673 | 0.05 |
ENST00000397473.2 ENST00000397470.2 |
GPR18 |
G protein-coupled receptor 18 |
| chr9_+_71944241 | 0.05 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr9_-_104145795 | 0.05 |
ENST00000259407.2 |
BAAT |
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr3_-_52486841 | 0.05 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr1_-_153283194 | 0.05 |
ENST00000290722.1 |
PGLYRP3 |
peptidoglycan recognition protein 3 |
| chr18_+_55862622 | 0.05 |
ENST00000456173.2 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr6_-_27835357 | 0.05 |
ENST00000331442.3 |
HIST1H1B |
histone cluster 1, H1b |
| chr13_+_24553933 | 0.05 |
ENST00000424834.2 ENST00000439928.2 |
SPATA13 RP11-309I15.1 |
spermatogenesis associated 13 RP11-309I15.1 |
| chr8_+_50824233 | 0.05 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr4_+_55095428 | 0.05 |
ENST00000508170.1 ENST00000512143.1 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr6_-_56507586 | 0.05 |
ENST00000439203.1 ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST |
dystonin |
| chr8_-_27115903 | 0.05 |
ENST00000350889.4 ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4 |
stathmin-like 4 |
| chr5_-_150603679 | 0.05 |
ENST00000355417.2 |
CCDC69 |
coiled-coil domain containing 69 |
| chrX_+_21392529 | 0.05 |
ENST00000425654.2 ENST00000543067.1 |
CNKSR2 |
connector enhancer of kinase suppressor of Ras 2 |
| chr1_+_16010779 | 0.05 |
ENST00000375799.3 ENST00000375793.2 |
PLEKHM2 |
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr7_+_37960163 | 0.05 |
ENST00000199448.4 ENST00000559325.1 ENST00000423717.1 |
EPDR1 |
ependymin related 1 |
| chr19_-_54876558 | 0.05 |
ENST00000391742.2 ENST00000434277.2 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
| chr14_-_23451845 | 0.05 |
ENST00000262713.2 |
AJUBA |
ajuba LIM protein |
| chr1_+_160765947 | 0.05 |
ENST00000263285.6 ENST00000368039.2 |
LY9 |
lymphocyte antigen 9 |
| chr12_+_86268065 | 0.05 |
ENST00000551529.1 ENST00000256010.6 |
NTS |
neurotensin |
| chr9_+_125281420 | 0.05 |
ENST00000340750.1 |
OR1J4 |
olfactory receptor, family 1, subfamily J, member 4 |
| chr4_-_155511887 | 0.05 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chr6_+_96025341 | 0.05 |
ENST00000369293.1 ENST00000358812.4 |
MANEA |
mannosidase, endo-alpha |
| chr11_-_71639670 | 0.05 |
ENST00000533047.1 ENST00000529844.1 |
RP11-849H4.2 |
Putative short transient receptor potential channel 2-like protein |
| chr13_-_38443860 | 0.05 |
ENST00000426868.2 ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4 |
transient receptor potential cation channel, subfamily C, member 4 |
| chr1_-_156252590 | 0.05 |
ENST00000361813.5 ENST00000368267.5 |
SMG5 |
SMG5 nonsense mediated mRNA decay factor |
| chr8_-_114389353 | 0.05 |
ENST00000343508.3 |
CSMD3 |
CUB and Sushi multiple domains 3 |
| chr8_-_70745575 | 0.05 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr7_-_43946175 | 0.05 |
ENST00000603700.1 ENST00000402306.3 ENST00000443736.1 ENST00000446958.1 |
URGCP-MRPS24 URGCP |
URGCP-MRPS24 readthrough upregulator of cell proliferation |
| chr13_+_78315466 | 0.04 |
ENST00000314070.5 ENST00000462234.1 |
SLAIN1 |
SLAIN motif family, member 1 |
| chr1_-_102462565 | 0.04 |
ENST00000370103.4 |
OLFM3 |
olfactomedin 3 |
| chr6_+_123317116 | 0.04 |
ENST00000275162.5 |
CLVS2 |
clavesin 2 |
| chr22_+_46449674 | 0.04 |
ENST00000381051.2 |
FLJ27365 |
hsa-mir-4763 |
| chr2_+_145425573 | 0.04 |
ENST00000600064.1 ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr17_-_29624343 | 0.04 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr8_+_67405794 | 0.04 |
ENST00000522977.1 ENST00000480005.1 |
C8orf46 |
chromosome 8 open reading frame 46 |
| chr22_-_46450024 | 0.04 |
ENST00000396008.2 ENST00000333761.1 |
C22orf26 |
chromosome 22 open reading frame 26 |
| chr2_-_105882585 | 0.04 |
ENST00000595531.1 |
AC012360.2 |
LOC644617 protein; Uncharacterized protein |
| chr1_+_169079823 | 0.04 |
ENST00000367813.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr3_-_119379719 | 0.04 |
ENST00000493094.1 |
POPDC2 |
popeye domain containing 2 |
| chr1_-_240906911 | 0.04 |
ENST00000431139.2 |
RP11-80B9.4 |
RP11-80B9.4 |
| chr5_+_57878859 | 0.04 |
ENST00000282878.4 |
RAB3C |
RAB3C, member RAS oncogene family |
| chr7_+_23719749 | 0.04 |
ENST00000409192.3 ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A |
family with sequence similarity 221, member A |
| chr14_+_56127989 | 0.04 |
ENST00000555573.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr8_+_65285851 | 0.04 |
ENST00000520799.1 ENST00000521441.1 |
LINC00966 |
long intergenic non-protein coding RNA 966 |
| chrX_-_54824673 | 0.04 |
ENST00000218436.6 |
ITIH6 |
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr19_+_19144384 | 0.04 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr17_+_57274914 | 0.04 |
ENST00000582004.1 ENST00000577660.1 |
PRR11 CTD-2510F5.6 |
proline rich 11 Uncharacterized protein |
| chr3_+_132316081 | 0.04 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr2_+_145425534 | 0.04 |
ENST00000432608.1 ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr11_-_71639480 | 0.04 |
ENST00000529513.1 |
RP11-849H4.2 |
Putative short transient receptor potential channel 2-like protein |
| chr2_-_136875712 | 0.04 |
ENST00000241393.3 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chr5_+_173472607 | 0.04 |
ENST00000303177.3 ENST00000519867.1 |
NSG2 |
Neuron-specific protein family member 2 |
| chr2_+_133874577 | 0.04 |
ENST00000596384.1 |
AC011755.1 |
HCG2006742; Protein LOC100996685 |
| chr1_+_109256067 | 0.04 |
ENST00000271311.2 |
FNDC7 |
fibronectin type III domain containing 7 |
| chr6_+_131958436 | 0.03 |
ENST00000357639.3 ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr10_-_61718429 | 0.03 |
ENST00000430888.1 |
C10orf40 |
chromosome 10 open reading frame 40 |
| chr7_+_77167343 | 0.03 |
ENST00000433369.2 ENST00000415482.2 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
| chr15_-_22448819 | 0.03 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr6_+_31638156 | 0.03 |
ENST00000409525.1 |
LY6G5B |
lymphocyte antigen 6 complex, locus G5B |
| chr10_+_50507181 | 0.03 |
ENST00000323868.4 |
C10orf71 |
chromosome 10 open reading frame 71 |
| chr15_+_80733570 | 0.03 |
ENST00000533983.1 ENST00000527771.1 ENST00000525103.1 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr18_+_3466248 | 0.03 |
ENST00000581029.1 ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4 |
RP11-838N2.4 |
| chr1_+_32716840 | 0.03 |
ENST00000336890.5 |
LCK |
lymphocyte-specific protein tyrosine kinase |
| chr17_+_7328925 | 0.03 |
ENST00000333870.3 ENST00000574034.1 |
C17orf74 |
chromosome 17 open reading frame 74 |
| chr3_-_33686925 | 0.03 |
ENST00000485378.2 ENST00000313350.6 ENST00000487200.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr21_-_27423339 | 0.03 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr10_+_50507232 | 0.03 |
ENST00000374144.3 |
C10orf71 |
chromosome 10 open reading frame 71 |
| chrX_-_10588459 | 0.03 |
ENST00000380782.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr13_-_95131923 | 0.03 |
ENST00000377028.5 ENST00000446125.1 |
DCT |
dopachrome tautomerase |
| chrX_-_55024967 | 0.03 |
ENST00000545676.1 |
PFKFB1 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr2_+_238767517 | 0.03 |
ENST00000404910.2 |
RAMP1 |
receptor (G protein-coupled) activity modifying protein 1 |
| chr3_-_11685345 | 0.03 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chrX_-_10588595 | 0.03 |
ENST00000423614.1 ENST00000317552.4 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr22_-_50523760 | 0.03 |
ENST00000395876.2 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr11_+_55029628 | 0.03 |
ENST00000417545.2 |
TRIM48 |
tripartite motif containing 48 |
| chr6_-_42690312 | 0.03 |
ENST00000230381.5 |
PRPH2 |
peripherin 2 (retinal degeneration, slow) |
| chr19_+_17862274 | 0.03 |
ENST00000596536.1 ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1 |
FCH domain only 1 |
| chr12_-_111806892 | 0.03 |
ENST00000547710.1 ENST00000549321.1 ENST00000361483.3 ENST00000392658.5 |
FAM109A |
family with sequence similarity 109, member A |
| chr2_-_151344172 | 0.03 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr17_+_47865917 | 0.03 |
ENST00000259021.4 ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7 |
K(lysine) acetyltransferase 7 |
| chr6_+_46761118 | 0.03 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr15_+_59730348 | 0.03 |
ENST00000288228.5 ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A |
family with sequence similarity 81, member A |
| chr7_+_116660246 | 0.03 |
ENST00000434836.1 ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr2_+_210636697 | 0.03 |
ENST00000439458.1 ENST00000272845.6 |
UNC80 |
unc-80 homolog (C. elegans) |
| chr16_+_33006369 | 0.03 |
ENST00000425181.3 |
IGHV3OR16-10 |
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr12_-_111358372 | 0.03 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chr6_-_32634425 | 0.03 |
ENST00000399082.3 ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1 |
major histocompatibility complex, class II, DQ beta 1 |
| chr4_+_41614909 | 0.02 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr6_+_109169591 | 0.02 |
ENST00000368972.3 ENST00000392644.4 |
ARMC2 |
armadillo repeat containing 2 |
| chr20_+_62716348 | 0.02 |
ENST00000349451.3 |
OPRL1 |
opiate receptor-like 1 |
| chr4_+_36283213 | 0.02 |
ENST00000357504.3 |
DTHD1 |
death domain containing 1 |
| chr9_+_90497741 | 0.02 |
ENST00000325643.5 |
SPATA31E1 |
SPATA31 subfamily E, member 1 |
| chr6_-_45983549 | 0.02 |
ENST00000544153.1 |
CLIC5 |
chloride intracellular channel 5 |
| chr16_-_20681177 | 0.02 |
ENST00000524149.1 |
ACSM1 |
acyl-CoA synthetase medium-chain family member 1 |
| chr11_-_89653576 | 0.02 |
ENST00000420869.1 |
TRIM49D1 |
tripartite motif containing 49D1 |
| chr21_-_33975547 | 0.02 |
ENST00000431599.1 |
C21orf59 |
chromosome 21 open reading frame 59 |
| chr3_+_130279178 | 0.02 |
ENST00000358511.6 ENST00000453409.2 |
COL6A6 |
collagen, type VI, alpha 6 |
| chr2_-_48468122 | 0.02 |
ENST00000447571.1 |
AC079807.4 |
AC079807.4 |
| chr21_-_44299626 | 0.02 |
ENST00000330317.2 ENST00000398208.2 |
WDR4 |
WD repeat domain 4 |
| chr1_-_87379785 | 0.02 |
ENST00000401030.3 ENST00000370554.1 |
SEP15 |
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr12_+_123011776 | 0.02 |
ENST00000450485.2 ENST00000333479.7 |
KNTC1 |
kinetochore associated 1 |
| chr14_+_68286478 | 0.02 |
ENST00000487861.1 |
RAD51B |
RAD51 paralog B |
| chr4_-_13546632 | 0.02 |
ENST00000382438.5 |
NKX3-2 |
NK3 homeobox 2 |
| chr19_+_9296279 | 0.02 |
ENST00000344248.2 |
OR7D2 |
olfactory receptor, family 7, subfamily D, member 2 |
| chr3_+_63428982 | 0.02 |
ENST00000479198.1 ENST00000460711.1 ENST00000465156.1 |
SYNPR |
synaptoporin |
| chr4_+_71296204 | 0.02 |
ENST00000413702.1 |
MUC7 |
mucin 7, secreted |
| chr10_+_123923205 | 0.02 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr2_+_38893047 | 0.02 |
ENST00000272252.5 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
| chrX_-_15402498 | 0.02 |
ENST00000297904.3 |
FIGF |
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr2_-_196933536 | 0.02 |
ENST00000312428.6 ENST00000410072.1 |
DNAH7 |
dynein, axonemal, heavy chain 7 |
| chr6_-_52705641 | 0.02 |
ENST00000370989.2 |
GSTA5 |
glutathione S-transferase alpha 5 |
| chr14_+_68286516 | 0.02 |
ENST00000471583.1 ENST00000487270.1 ENST00000488612.1 |
RAD51B |
RAD51 paralog B |
| chr7_+_123488124 | 0.02 |
ENST00000476325.1 |
HYAL4 |
hyaluronoglucosaminidase 4 |
| chr20_-_23402028 | 0.02 |
ENST00000398425.3 ENST00000432543.2 ENST00000377026.4 |
NAPB |
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr7_+_54610124 | 0.02 |
ENST00000402026.2 |
VSTM2A |
V-set and transmembrane domain containing 2A |
| chr22_-_50524298 | 0.02 |
ENST00000311597.5 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr17_-_39306054 | 0.02 |
ENST00000343246.4 |
KRTAP4-5 |
keratin associated protein 4-5 |
| chr3_-_25915189 | 0.02 |
ENST00000451284.1 |
LINC00692 |
long intergenic non-protein coding RNA 692 |
| chr11_+_49050504 | 0.01 |
ENST00000332682.7 |
TRIM49B |
tripartite motif containing 49B |
| chr1_-_39339777 | 0.01 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chr13_+_28813645 | 0.01 |
ENST00000282391.5 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr1_+_171283331 | 0.01 |
ENST00000367749.3 |
FMO4 |
flavin containing monooxygenase 4 |
| chr5_-_108063949 | 0.01 |
ENST00000606054.1 |
LINC01023 |
long intergenic non-protein coding RNA 1023 |
| chr6_-_29399744 | 0.01 |
ENST00000377154.1 |
OR5V1 |
olfactory receptor, family 5, subfamily V, member 1 |
| chr2_+_108905325 | 0.01 |
ENST00000438339.1 ENST00000409880.1 ENST00000437390.2 |
SULT1C2 |
sulfotransferase family, cytosolic, 1C, member 2 |
| chr7_-_36634181 | 0.01 |
ENST00000538464.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 1.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.3 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.2 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0051714 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 1.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |