Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
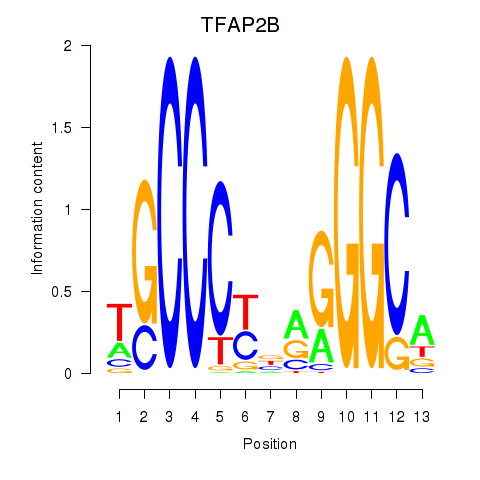
Results for TFAP2B
Z-value: 1.91
Transcription factors associated with TFAP2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2B
|
ENSG00000008196.8 | TFAP2B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP2B | hg19_v2_chr6_+_50786414_50786439 | 0.54 | 1.7e-01 | Click! |
Activity profile of TFAP2B motif
Sorted Z-values of TFAP2B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2B
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39677971 | 2.45 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr1_+_160370344 | 2.02 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr16_+_68679193 | 1.95 |
ENST00000581171.1 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr1_-_153588334 | 1.91 |
ENST00000476873.1 |
S100A14 |
S100 calcium binding protein A14 |
| chr16_+_22825475 | 1.87 |
ENST00000261374.3 |
HS3ST2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr1_+_209602771 | 1.74 |
ENST00000440276.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr16_+_68678892 | 1.74 |
ENST00000429102.2 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr1_+_209602609 | 1.63 |
ENST00000458250.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr19_-_19739007 | 1.55 |
ENST00000586703.1 ENST00000591042.1 ENST00000407877.3 |
LPAR2 |
lysophosphatidic acid receptor 2 |
| chr15_+_101420028 | 1.55 |
ENST00000557963.1 ENST00000346623.6 |
ALDH1A3 |
aldehyde dehydrogenase 1 family, member A3 |
| chr14_+_65171315 | 1.40 |
ENST00000394691.1 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr2_+_120770686 | 1.38 |
ENST00000331393.4 ENST00000443124.1 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr8_-_21988558 | 1.33 |
ENST00000312841.8 |
HR |
hair growth associated |
| chr6_-_136871957 | 1.24 |
ENST00000354570.3 |
MAP7 |
microtubule-associated protein 7 |
| chr20_+_62327996 | 1.22 |
ENST00000369996.1 |
TNFRSF6B |
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr7_-_143105941 | 1.20 |
ENST00000275815.3 |
EPHA1 |
EPH receptor A1 |
| chr6_+_54711533 | 1.20 |
ENST00000306858.7 |
FAM83B |
family with sequence similarity 83, member B |
| chr18_+_33877654 | 1.19 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr16_+_81478775 | 1.07 |
ENST00000537098.3 |
CMIP |
c-Maf inducing protein |
| chr19_-_55660561 | 1.06 |
ENST00000587758.1 ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr7_+_16793160 | 1.06 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr16_-_68269971 | 1.05 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr16_+_67465016 | 0.97 |
ENST00000326152.5 |
HSD11B2 |
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr1_+_2036149 | 0.97 |
ENST00000482686.1 ENST00000400920.1 ENST00000486681.1 |
PRKCZ |
protein kinase C, zeta |
| chr19_+_7710774 | 0.95 |
ENST00000602355.1 |
STXBP2 |
syntaxin binding protein 2 |
| chr1_+_1981890 | 0.92 |
ENST00000378567.3 ENST00000468310.1 |
PRKCZ |
protein kinase C, zeta |
| chr1_+_233463507 | 0.86 |
ENST00000366623.3 ENST00000366624.3 |
MLK4 |
Mitogen-activated protein kinase kinase kinase MLK4 |
| chr1_+_65613852 | 0.83 |
ENST00000327299.7 |
AK4 |
adenylate kinase 4 |
| chr14_+_65171099 | 0.83 |
ENST00000247226.7 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr1_+_32042105 | 0.82 |
ENST00000457433.2 ENST00000441210.2 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
| chr14_+_94640633 | 0.81 |
ENST00000304338.3 |
PPP4R4 |
protein phosphatase 4, regulatory subunit 4 |
| chr22_-_20255212 | 0.81 |
ENST00000416372.1 |
RTN4R |
reticulon 4 receptor |
| chr6_-_138428613 | 0.79 |
ENST00000421351.3 |
PERP |
PERP, TP53 apoptosis effector |
| chr1_+_32042131 | 0.79 |
ENST00000271064.7 ENST00000537531.1 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
| chr1_-_27286897 | 0.75 |
ENST00000320567.5 |
C1orf172 |
chromosome 1 open reading frame 172 |
| chr5_+_36608422 | 0.75 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr16_+_3014217 | 0.72 |
ENST00000572045.1 |
KREMEN2 |
kringle containing transmembrane protein 2 |
| chr1_-_9189229 | 0.71 |
ENST00000377411.4 |
GPR157 |
G protein-coupled receptor 157 |
| chr1_+_152881014 | 0.71 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr12_-_89746173 | 0.70 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr20_-_49639631 | 0.68 |
ENST00000424171.1 ENST00000439216.1 ENST00000371571.4 |
KCNG1 |
potassium voltage-gated channel, subfamily G, member 1 |
| chr20_-_50808236 | 0.62 |
ENST00000361387.2 |
ZFP64 |
ZFP64 zinc finger protein |
| chr22_+_45072925 | 0.62 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr16_+_2867228 | 0.62 |
ENST00000005995.3 ENST00000574813.1 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr22_+_45072958 | 0.62 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr19_+_7701985 | 0.61 |
ENST00000595950.1 ENST00000441779.2 ENST00000221283.5 ENST00000414284.2 |
STXBP2 |
syntaxin binding protein 2 |
| chr2_-_220408430 | 0.60 |
ENST00000243776.6 |
CHPF |
chondroitin polymerizing factor |
| chr17_-_4463856 | 0.60 |
ENST00000574584.1 ENST00000381550.3 ENST00000301395.3 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr10_+_48355024 | 0.59 |
ENST00000395702.2 ENST00000442001.1 ENST00000433077.1 ENST00000436850.1 ENST00000494156.1 ENST00000586537.1 |
ZNF488 |
zinc finger protein 488 |
| chr11_-_7695437 | 0.59 |
ENST00000533558.1 ENST00000527542.1 ENST00000531096.1 |
CYB5R2 |
cytochrome b5 reductase 2 |
| chr14_+_92980111 | 0.59 |
ENST00000216487.7 ENST00000557762.1 |
RIN3 |
Ras and Rab interactor 3 |
| chr3_-_48470838 | 0.58 |
ENST00000358459.4 ENST00000358536.4 |
PLXNB1 |
plexin B1 |
| chr11_-_7694684 | 0.58 |
ENST00000524790.1 ENST00000299497.9 ENST00000299498.6 |
CYB5R2 |
cytochrome b5 reductase 2 |
| chrX_-_78622805 | 0.57 |
ENST00000373298.2 |
ITM2A |
integral membrane protein 2A |
| chr16_+_2867164 | 0.57 |
ENST00000455114.1 ENST00000450020.3 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr5_+_131409476 | 0.57 |
ENST00000296871.2 |
CSF2 |
colony stimulating factor 2 (granulocyte-macrophage) |
| chr1_-_23886285 | 0.56 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr3_-_196756646 | 0.56 |
ENST00000439320.1 ENST00000296351.4 ENST00000296350.5 |
MFI2 |
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| chr8_-_144655141 | 0.56 |
ENST00000398882.3 |
MROH6 |
maestro heat-like repeat family member 6 |
| chr13_-_76056250 | 0.54 |
ENST00000377636.3 ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4 |
TBC1 domain family, member 4 |
| chr5_-_150603679 | 0.54 |
ENST00000355417.2 |
CCDC69 |
coiled-coil domain containing 69 |
| chr3_-_185216811 | 0.51 |
ENST00000421852.1 |
TMEM41A |
transmembrane protein 41A |
| chr1_+_33207381 | 0.51 |
ENST00000401073.2 |
KIAA1522 |
KIAA1522 |
| chr1_+_153746683 | 0.51 |
ENST00000271857.2 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr19_+_17858509 | 0.50 |
ENST00000594202.1 ENST00000252771.7 ENST00000389133.4 |
FCHO1 |
FCH domain only 1 |
| chr19_+_17858547 | 0.50 |
ENST00000600676.1 ENST00000600209.1 ENST00000596309.1 ENST00000598539.1 ENST00000597474.1 ENST00000593385.1 ENST00000598067.1 ENST00000593833.1 |
FCHO1 |
FCH domain only 1 |
| chr3_-_52090461 | 0.50 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr8_+_123793633 | 0.50 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr9_+_116917807 | 0.49 |
ENST00000356083.3 |
COL27A1 |
collagen, type XXVII, alpha 1 |
| chr22_-_46933067 | 0.49 |
ENST00000262738.3 ENST00000395964.1 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr21_+_18885318 | 0.49 |
ENST00000400166.1 |
CXADR |
coxsackie virus and adenovirus receptor |
| chr3_-_124839648 | 0.49 |
ENST00000430155.2 |
SLC12A8 |
solute carrier family 12, member 8 |
| chr19_-_11689752 | 0.48 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr12_-_31477072 | 0.48 |
ENST00000454658.2 |
FAM60A |
family with sequence similarity 60, member A |
| chr2_-_73511559 | 0.47 |
ENST00000521871.1 |
FBXO41 |
F-box protein 41 |
| chr9_+_133971909 | 0.47 |
ENST00000247291.3 ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L |
allograft inflammatory factor 1-like |
| chr16_+_3014269 | 0.46 |
ENST00000575885.1 ENST00000571007.1 ENST00000319500.6 |
KREMEN2 |
kringle containing transmembrane protein 2 |
| chr9_+_133971863 | 0.46 |
ENST00000372309.3 |
AIF1L |
allograft inflammatory factor 1-like |
| chr8_+_56792377 | 0.46 |
ENST00000520220.2 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr8_+_56792355 | 0.46 |
ENST00000519728.1 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr17_-_39942940 | 0.46 |
ENST00000310706.5 ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP |
junction plakoglobin |
| chr12_+_7060432 | 0.43 |
ENST00000318974.9 ENST00000456013.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr1_-_31902614 | 0.43 |
ENST00000596131.1 |
AC114494.1 |
HCG1787699; Uncharacterized protein |
| chr16_+_23847267 | 0.43 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr12_-_102224457 | 0.43 |
ENST00000549165.1 ENST00000549940.1 ENST00000392919.4 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr1_-_147245484 | 0.42 |
ENST00000271348.2 |
GJA5 |
gap junction protein, alpha 5, 40kDa |
| chr12_+_6493406 | 0.42 |
ENST00000543190.1 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr6_-_3457256 | 0.42 |
ENST00000436008.2 |
SLC22A23 |
solute carrier family 22, member 23 |
| chr1_+_55013889 | 0.41 |
ENST00000343744.2 ENST00000371316.3 |
ACOT11 |
acyl-CoA thioesterase 11 |
| chr19_+_1067492 | 0.41 |
ENST00000586866.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr19_+_1067144 | 0.41 |
ENST00000313093.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr19_+_1067271 | 0.40 |
ENST00000536472.1 ENST00000590214.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr5_-_174871136 | 0.40 |
ENST00000393752.2 |
DRD1 |
dopamine receptor D1 |
| chr1_-_19283163 | 0.40 |
ENST00000455833.2 |
IFFO2 |
intermediate filament family orphan 2 |
| chr19_-_42724261 | 0.39 |
ENST00000595337.1 |
DEDD2 |
death effector domain containing 2 |
| chr1_-_54872059 | 0.39 |
ENST00000371320.3 |
SSBP3 |
single stranded DNA binding protein 3 |
| chr12_+_6308881 | 0.38 |
ENST00000382518.1 ENST00000536586.1 |
CD9 |
CD9 molecule |
| chr17_+_1646130 | 0.38 |
ENST00000453066.1 ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr17_-_73874654 | 0.38 |
ENST00000254816.2 |
TRIM47 |
tripartite motif containing 47 |
| chr5_+_149737202 | 0.38 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
| chr1_+_45792541 | 0.38 |
ENST00000334815.3 |
HPDL |
4-hydroxyphenylpyruvate dioxygenase-like |
| chr1_-_25291475 | 0.38 |
ENST00000338888.3 ENST00000399916.1 |
RUNX3 |
runt-related transcription factor 3 |
| chr3_+_10857885 | 0.36 |
ENST00000254488.2 ENST00000454147.1 |
SLC6A11 |
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chrX_+_37545012 | 0.35 |
ENST00000378616.3 |
XK |
X-linked Kx blood group (McLeod syndrome) |
| chrX_+_21392873 | 0.35 |
ENST00000379510.3 |
CNKSR2 |
connector enhancer of kinase suppressor of Ras 2 |
| chr11_+_63606558 | 0.34 |
ENST00000350490.7 ENST00000502399.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr13_-_99852916 | 0.34 |
ENST00000426037.2 ENST00000445737.2 |
UBAC2-AS1 |
UBAC2 antisense RNA 1 |
| chr16_+_81678957 | 0.34 |
ENST00000398040.4 |
CMIP |
c-Maf inducing protein |
| chr4_-_1723040 | 0.33 |
ENST00000382936.3 ENST00000536901.1 ENST00000303277.2 |
TMEM129 |
transmembrane protein 129 |
| chr1_-_15911510 | 0.33 |
ENST00000375826.3 |
AGMAT |
agmatine ureohydrolase (agmatinase) |
| chr15_-_79383102 | 0.32 |
ENST00000558480.2 ENST00000419573.3 |
RASGRF1 |
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr3_-_185216766 | 0.32 |
ENST00000296254.3 |
TMEM41A |
transmembrane protein 41A |
| chr2_-_208030647 | 0.31 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr2_+_26915584 | 0.31 |
ENST00000302909.3 |
KCNK3 |
potassium channel, subfamily K, member 3 |
| chr12_+_50451462 | 0.31 |
ENST00000447966.2 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chr2_-_208634287 | 0.31 |
ENST00000295417.3 |
FZD5 |
frizzled family receptor 5 |
| chrX_+_48916497 | 0.30 |
ENST00000496529.2 ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120 |
coiled-coil domain containing 120 |
| chr1_+_32716840 | 0.30 |
ENST00000336890.5 |
LCK |
lymphocyte-specific protein tyrosine kinase |
| chr3_+_125687987 | 0.30 |
ENST00000514116.1 ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr8_-_8751068 | 0.29 |
ENST00000276282.6 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
| chr6_-_35888905 | 0.29 |
ENST00000510290.1 ENST00000423325.2 ENST00000373822.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr10_-_35930219 | 0.29 |
ENST00000374694.1 |
FZD8 |
frizzled family receptor 8 |
| chrX_+_131157290 | 0.29 |
ENST00000394334.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr17_-_7760779 | 0.29 |
ENST00000335155.5 ENST00000575071.1 |
LSMD1 |
LSM domain containing 1 |
| chr7_-_151329416 | 0.28 |
ENST00000418337.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr19_-_49149553 | 0.28 |
ENST00000084798.4 |
CA11 |
carbonic anhydrase XI |
| chrX_-_154033793 | 0.28 |
ENST00000369534.3 ENST00000413259.3 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chr1_+_155178518 | 0.28 |
ENST00000316721.4 |
MTX1 |
metaxin 1 |
| chr10_-_99094458 | 0.27 |
ENST00000371019.2 |
FRAT2 |
frequently rearranged in advanced T-cell lymphomas 2 |
| chr12_-_122238913 | 0.27 |
ENST00000537157.1 |
AC084018.1 |
AC084018.1 |
| chr6_+_33589161 | 0.27 |
ENST00000605930.1 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr12_+_19282713 | 0.27 |
ENST00000299275.6 ENST00000539256.1 ENST00000538714.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr10_+_103825080 | 0.27 |
ENST00000299238.5 |
HPS6 |
Hermansky-Pudlak syndrome 6 |
| chr19_+_39989580 | 0.26 |
ENST00000596614.1 ENST00000205143.4 |
DLL3 |
delta-like 3 (Drosophila) |
| chrX_+_131157322 | 0.26 |
ENST00000481105.1 ENST00000354719.6 ENST00000394335.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr6_-_35888824 | 0.26 |
ENST00000361690.3 ENST00000512445.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr19_+_30433372 | 0.26 |
ENST00000312051.6 |
URI1 |
URI1, prefoldin-like chaperone |
| chr8_-_143867946 | 0.25 |
ENST00000301263.4 |
LY6D |
lymphocyte antigen 6 complex, locus D |
| chr5_+_141016969 | 0.25 |
ENST00000518856.1 |
RELL2 |
RELT-like 2 |
| chr12_+_56414851 | 0.25 |
ENST00000547167.1 |
IKZF4 |
IKAROS family zinc finger 4 (Eos) |
| chr1_+_17575584 | 0.25 |
ENST00000375460.3 |
PADI3 |
peptidyl arginine deiminase, type III |
| chr19_+_19144384 | 0.25 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr21_-_47575481 | 0.25 |
ENST00000291670.5 ENST00000397748.1 ENST00000359679.2 ENST00000355384.2 ENST00000397746.3 ENST00000397743.1 |
FTCD |
formimidoyltransferase cyclodeaminase |
| chr3_-_150481164 | 0.25 |
ENST00000312960.3 |
SIAH2 |
siah E3 ubiquitin protein ligase 2 |
| chr14_+_94640671 | 0.24 |
ENST00000328839.3 |
PPP4R4 |
protein phosphatase 4, regulatory subunit 4 |
| chr6_-_47277634 | 0.24 |
ENST00000296861.2 |
TNFRSF21 |
tumor necrosis factor receptor superfamily, member 21 |
| chr2_+_101869262 | 0.23 |
ENST00000289382.3 |
CNOT11 |
CCR4-NOT transcription complex, subunit 11 |
| chr4_-_84030996 | 0.23 |
ENST00000411416.2 |
PLAC8 |
placenta-specific 8 |
| chr6_+_143381979 | 0.23 |
ENST00000367598.5 ENST00000447498.1 ENST00000357847.4 ENST00000344492.5 ENST00000367596.1 ENST00000494282.2 ENST00000275235.4 |
AIG1 |
androgen-induced 1 |
| chr19_+_38397839 | 0.23 |
ENST00000222345.6 |
SIPA1L3 |
signal-induced proliferation-associated 1 like 3 |
| chr1_+_42921761 | 0.23 |
ENST00000372562.1 |
PPCS |
phosphopantothenoylcysteine synthetase |
| chr9_-_124976185 | 0.23 |
ENST00000464484.2 |
LHX6 |
LIM homeobox 6 |
| chr5_+_141016508 | 0.23 |
ENST00000444782.1 ENST00000521367.1 ENST00000297164.3 |
RELL2 |
RELT-like 2 |
| chr5_-_134369973 | 0.22 |
ENST00000265340.7 |
PITX1 |
paired-like homeodomain 1 |
| chr7_-_150498426 | 0.22 |
ENST00000447204.2 |
TMEM176B |
transmembrane protein 176B |
| chrX_+_131157609 | 0.22 |
ENST00000496850.1 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr17_-_43502987 | 0.22 |
ENST00000376922.2 |
ARHGAP27 |
Rho GTPase activating protein 27 |
| chr9_-_124976154 | 0.22 |
ENST00000482062.1 |
LHX6 |
LIM homeobox 6 |
| chr19_+_39989535 | 0.22 |
ENST00000356433.5 |
DLL3 |
delta-like 3 (Drosophila) |
| chr16_+_3194211 | 0.22 |
ENST00000428155.1 |
CASP16 |
caspase 16, apoptosis-related cysteine peptidase (putative) |
| chrX_+_70316005 | 0.22 |
ENST00000374259.3 |
FOXO4 |
forkhead box O4 |
| chr1_+_155179012 | 0.21 |
ENST00000609421.1 |
MTX1 |
metaxin 1 |
| chr15_+_90319557 | 0.21 |
ENST00000341735.3 |
MESP2 |
mesoderm posterior 2 homolog (mouse) |
| chr14_+_24540046 | 0.21 |
ENST00000397016.2 ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6 |
copine VI (neuronal) |
| chr1_-_155006224 | 0.21 |
ENST00000368424.3 |
DCST2 |
DC-STAMP domain containing 2 |
| chr1_+_107683436 | 0.21 |
ENST00000370068.1 |
NTNG1 |
netrin G1 |
| chr10_-_8095412 | 0.21 |
ENST00000458727.1 ENST00000355358.1 |
RP11-379F12.3 GATA3-AS1 |
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr12_+_6493319 | 0.21 |
ENST00000536876.1 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr19_+_42301079 | 0.21 |
ENST00000596544.1 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr19_+_42387228 | 0.21 |
ENST00000354532.3 ENST00000599846.1 ENST00000347545.4 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr5_-_93077293 | 0.21 |
ENST00000510627.4 |
POU5F2 |
POU domain class 5, transcription factor 2 |
| chr17_+_79761997 | 0.21 |
ENST00000400723.3 ENST00000570996.1 |
GCGR |
glucagon receptor |
| chr1_+_29213584 | 0.20 |
ENST00000343067.4 ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr16_-_66959429 | 0.20 |
ENST00000420652.1 ENST00000299759.6 |
RRAD |
Ras-related associated with diabetes |
| chr1_+_107683644 | 0.20 |
ENST00000370067.1 |
NTNG1 |
netrin G1 |
| chr12_-_15114492 | 0.20 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr5_+_61708582 | 0.20 |
ENST00000325324.6 |
IPO11 |
importin 11 |
| chr3_-_48471454 | 0.20 |
ENST00000296440.6 ENST00000448774.2 |
PLXNB1 |
plexin B1 |
| chr13_+_52158610 | 0.20 |
ENST00000298125.5 |
WDFY2 |
WD repeat and FYVE domain containing 2 |
| chr1_-_6321035 | 0.20 |
ENST00000377893.2 |
GPR153 |
G protein-coupled receptor 153 |
| chr3_-_52443799 | 0.20 |
ENST00000470173.1 ENST00000296288.5 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr2_-_191399426 | 0.19 |
ENST00000409150.3 |
TMEM194B |
transmembrane protein 194B |
| chr22_+_21987005 | 0.19 |
ENST00000607942.1 ENST00000425975.1 ENST00000292779.3 |
CCDC116 |
coiled-coil domain containing 116 |
| chr1_+_155178481 | 0.19 |
ENST00000368376.3 |
MTX1 |
metaxin 1 |
| chr13_-_25745857 | 0.19 |
ENST00000381853.3 |
AMER2 |
APC membrane recruitment protein 2 |
| chr21_+_37692481 | 0.19 |
ENST00000400485.1 |
MORC3 |
MORC family CW-type zinc finger 3 |
| chr6_-_34664612 | 0.19 |
ENST00000374023.3 ENST00000374026.3 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chrX_+_21392529 | 0.19 |
ENST00000425654.2 ENST00000543067.1 |
CNKSR2 |
connector enhancer of kinase suppressor of Ras 2 |
| chr2_+_112812778 | 0.19 |
ENST00000283206.4 |
TMEM87B |
transmembrane protein 87B |
| chr11_-_64512469 | 0.19 |
ENST00000377485.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr12_-_15114191 | 0.19 |
ENST00000541380.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr19_+_13229126 | 0.19 |
ENST00000292431.4 |
NACC1 |
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr20_-_49639612 | 0.18 |
ENST00000396017.3 ENST00000433903.1 |
KCNG1 |
potassium voltage-gated channel, subfamily G, member 1 |
| chr12_-_15114603 | 0.18 |
ENST00000228945.4 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr19_+_35630022 | 0.18 |
ENST00000589209.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr19_-_1650666 | 0.18 |
ENST00000588136.1 |
TCF3 |
transcription factor 3 |
| chr12_-_95941987 | 0.18 |
ENST00000537435.2 |
USP44 |
ubiquitin specific peptidase 44 |
| chr15_-_50647370 | 0.18 |
ENST00000558970.1 ENST00000396464.3 ENST00000560825.1 |
GABPB1 |
GA binding protein transcription factor, beta subunit 1 |
| chr17_-_17399701 | 0.18 |
ENST00000225688.3 ENST00000579152.1 |
RASD1 |
RAS, dexamethasone-induced 1 |
| chr4_+_78078304 | 0.18 |
ENST00000316355.5 ENST00000354403.5 ENST00000502280.1 |
CCNG2 |
cyclin G2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.8 | 2.5 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.5 | 1.5 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 0.9 | GO:0070666 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.3 | 1.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.3 | 0.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 0.7 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 0.7 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.6 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.2 | 0.9 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.2 | 0.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.4 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.6 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.6 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 1.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.7 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.3 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.8 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.8 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.2 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 1.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.5 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.9 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.3 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.3 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.5 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 0.2 | GO:0060066 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) oviduct development(GO:0060066) renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 | 1.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.3 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.8 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.3 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 1.0 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.4 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:1905176 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 1.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 1.7 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.7 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.9 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 1.2 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.5 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 1.0 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.6 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 2.9 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.3 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:1903276 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0015942 | formate metabolic process(GO:0015942) |
| 0.0 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.6 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.1 | GO:0072125 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) negative regulation of glomerulus development(GO:0090194) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 2.5 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0014063 | regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.0 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.0 | GO:0046666 | retinal cell programmed cell death(GO:0046666) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.3 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.2 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.9 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.2 | 1.5 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.6 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 1.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 1.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 4.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 1.0 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.3 | 1.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 0.8 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.3 | 1.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 1.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 1.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.2 | 0.8 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 1.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.4 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 1.9 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 2.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.2 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 1.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.9 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0031691 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 1.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |