Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
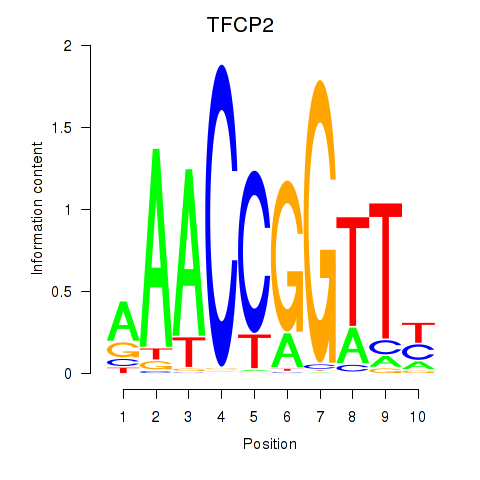
Results for TFCP2
Z-value: 0.16
Transcription factors associated with TFCP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2
|
ENSG00000135457.5 | TFCP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2 | hg19_v2_chr12_-_51566849_51566927 | 0.13 | 7.6e-01 | Click! |
Activity profile of TFCP2 motif
Sorted Z-values of TFCP2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_113542063 | 0.25 |
ENST00000263339.3 |
IL1A |
interleukin 1, alpha |
| chr11_+_62649158 | 0.25 |
ENST00000539891.1 ENST00000536981.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chrX_+_105937068 | 0.24 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_35247859 | 0.24 |
ENST00000373362.3 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr16_+_57653989 | 0.23 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr17_+_54671047 | 0.23 |
ENST00000332822.4 |
NOG |
noggin |
| chr12_+_41086297 | 0.22 |
ENST00000551295.2 |
CNTN1 |
contactin 1 |
| chr4_-_80994619 | 0.19 |
ENST00000404191.1 |
ANTXR2 |
anthrax toxin receptor 2 |
| chr1_+_86889769 | 0.17 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr10_-_90611566 | 0.17 |
ENST00000371930.4 |
ANKRD22 |
ankyrin repeat domain 22 |
| chr10_+_13203543 | 0.16 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chr7_+_127233689 | 0.16 |
ENST00000265825.5 ENST00000420086.2 |
FSCN3 |
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus) |
| chr2_+_189156389 | 0.16 |
ENST00000409843.1 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr4_-_89080003 | 0.16 |
ENST00000237612.3 |
ABCG2 |
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr12_-_51422017 | 0.15 |
ENST00000394904.3 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr1_-_54304212 | 0.15 |
ENST00000540001.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr2_-_61765315 | 0.14 |
ENST00000406957.1 ENST00000401558.2 |
XPO1 |
exportin 1 (CRM1 homolog, yeast) |
| chr1_+_109656532 | 0.14 |
ENST00000531664.1 ENST00000534476.1 |
KIAA1324 |
KIAA1324 |
| chr8_-_72459885 | 0.14 |
ENST00000523987.1 |
RP11-1102P16.1 |
Uncharacterized protein |
| chr4_-_174255400 | 0.13 |
ENST00000506267.1 |
HMGB2 |
high mobility group box 2 |
| chr17_+_47210125 | 0.13 |
ENST00000393354.2 |
B4GALNT2 |
beta-1,4-N-acetyl-galactosaminyl transferase 2 |
| chr16_+_838614 | 0.12 |
ENST00000262315.9 ENST00000455171.2 |
CHTF18 |
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr12_+_5153085 | 0.12 |
ENST00000252321.3 |
KCNA5 |
potassium voltage-gated channel, shaker-related subfamily, member 5 |
| chr4_+_128702969 | 0.11 |
ENST00000508776.1 ENST00000439123.2 |
HSPA4L |
heat shock 70kDa protein 4-like |
| chr7_-_105162652 | 0.10 |
ENST00000356362.2 ENST00000469408.1 |
PUS7 |
pseudouridylate synthase 7 homolog (S. cerevisiae) |
| chr3_+_184032919 | 0.10 |
ENST00000427845.1 ENST00000342981.4 ENST00000319274.6 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr6_+_142623063 | 0.10 |
ENST00000296932.8 ENST00000367609.3 |
GPR126 |
G protein-coupled receptor 126 |
| chr5_+_57878859 | 0.10 |
ENST00000282878.4 |
RAB3C |
RAB3C, member RAS oncogene family |
| chr10_+_6625605 | 0.10 |
ENST00000414894.1 ENST00000449648.1 |
PRKCQ-AS1 |
PRKCQ antisense RNA 1 |
| chr17_-_34308524 | 0.10 |
ENST00000293275.3 |
CCL16 |
chemokine (C-C motif) ligand 16 |
| chr12_+_41086215 | 0.09 |
ENST00000547702.1 ENST00000551424.1 |
CNTN1 |
contactin 1 |
| chr3_+_184033135 | 0.09 |
ENST00000424196.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_153330322 | 0.09 |
ENST00000368738.3 |
S100A9 |
S100 calcium binding protein A9 |
| chr12_+_122356488 | 0.09 |
ENST00000397454.2 |
WDR66 |
WD repeat domain 66 |
| chrX_+_151903253 | 0.09 |
ENST00000452779.2 ENST00000370291.2 |
CSAG1 |
chondrosarcoma associated gene 1 |
| chr10_-_76995675 | 0.09 |
ENST00000469299.1 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr11_-_62368696 | 0.09 |
ENST00000527204.1 |
MTA2 |
metastasis associated 1 family, member 2 |
| chr20_-_46415341 | 0.09 |
ENST00000484875.1 ENST00000361612.4 |
SULF2 |
sulfatase 2 |
| chr20_-_46415297 | 0.08 |
ENST00000467815.1 ENST00000359930.4 |
SULF2 |
sulfatase 2 |
| chr10_-_76995769 | 0.08 |
ENST00000372538.3 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr12_-_86230315 | 0.08 |
ENST00000361228.3 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chrX_+_151903207 | 0.08 |
ENST00000370287.3 |
CSAG1 |
chondrosarcoma associated gene 1 |
| chr4_-_166034029 | 0.08 |
ENST00000306480.6 |
TMEM192 |
transmembrane protein 192 |
| chr1_-_36947120 | 0.08 |
ENST00000361632.4 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
| chr8_-_17533838 | 0.08 |
ENST00000400046.1 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chrX_-_151903184 | 0.08 |
ENST00000357916.4 ENST00000393869.3 |
MAGEA12 |
melanoma antigen family A, 12 |
| chr3_-_183966717 | 0.07 |
ENST00000446569.1 ENST00000418734.2 ENST00000397676.3 |
ALG3 |
ALG3, alpha-1,3- mannosyltransferase |
| chr14_+_96722539 | 0.07 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chrX_-_151903101 | 0.07 |
ENST00000393900.3 |
MAGEA12 |
melanoma antigen family A, 12 |
| chr11_+_65554493 | 0.07 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr11_-_28129656 | 0.07 |
ENST00000263181.6 |
KIF18A |
kinesin family member 18A |
| chr4_+_170581213 | 0.07 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr2_+_63277927 | 0.07 |
ENST00000282549.2 |
OTX1 |
orthodenticle homeobox 1 |
| chrX_+_105066524 | 0.07 |
ENST00000243300.9 ENST00000428173.2 |
NRK |
Nik related kinase |
| chr1_-_153513170 | 0.07 |
ENST00000368717.2 |
S100A5 |
S100 calcium binding protein A5 |
| chr3_+_101568349 | 0.07 |
ENST00000326151.5 ENST00000326172.5 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr17_-_18430160 | 0.07 |
ENST00000392176.3 |
FAM106A |
family with sequence similarity 106, member A |
| chr11_+_6866883 | 0.07 |
ENST00000299454.4 ENST00000379831.2 |
OR10A5 |
olfactory receptor, family 10, subfamily A, member 5 |
| chr5_-_37371163 | 0.07 |
ENST00000513532.1 |
NUP155 |
nucleoporin 155kDa |
| chr1_-_246729544 | 0.07 |
ENST00000544618.1 ENST00000366514.4 |
TFB2M |
transcription factor B2, mitochondrial |
| chrX_-_72095622 | 0.07 |
ENST00000290273.5 |
DMRTC1 |
DMRT-like family C1 |
| chr11_+_113185292 | 0.06 |
ENST00000429951.1 ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12 |
tetratricopeptide repeat domain 12 |
| chr5_-_37371278 | 0.06 |
ENST00000231498.3 |
NUP155 |
nucleoporin 155kDa |
| chrX_-_72095808 | 0.06 |
ENST00000373529.5 |
DMRTC1 |
DMRT-like family C1 |
| chr17_-_5322786 | 0.06 |
ENST00000225696.4 |
NUP88 |
nucleoporin 88kDa |
| chr4_-_174255536 | 0.06 |
ENST00000446922.2 |
HMGB2 |
high mobility group box 2 |
| chr3_-_142297668 | 0.06 |
ENST00000350721.4 ENST00000383101.3 |
ATR |
ataxia telangiectasia and Rad3 related |
| chr11_-_128737163 | 0.06 |
ENST00000324003.3 ENST00000392665.2 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr18_-_67873078 | 0.06 |
ENST00000255674.6 |
RTTN |
rotatin |
| chr11_-_128737259 | 0.06 |
ENST00000440599.2 ENST00000392666.1 ENST00000324036.3 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr1_+_45212051 | 0.06 |
ENST00000372222.3 |
KIF2C |
kinesin family member 2C |
| chr5_+_110427983 | 0.06 |
ENST00000513710.2 ENST00000505303.1 |
WDR36 |
WD repeat domain 36 |
| chr1_-_41131326 | 0.06 |
ENST00000372684.3 |
RIMS3 |
regulating synaptic membrane exocytosis 3 |
| chr6_+_31371337 | 0.06 |
ENST00000449934.2 ENST00000421350.1 |
MICA |
MHC class I polypeptide-related sequence A |
| chr1_+_45212074 | 0.06 |
ENST00000372217.1 |
KIF2C |
kinesin family member 2C |
| chr5_+_61708582 | 0.06 |
ENST00000325324.6 |
IPO11 |
importin 11 |
| chr1_-_212588157 | 0.06 |
ENST00000261455.4 ENST00000535273.1 |
TMEM206 |
transmembrane protein 206 |
| chr16_-_82045049 | 0.06 |
ENST00000532128.1 ENST00000328945.5 |
SDR42E1 |
short chain dehydrogenase/reductase family 42E, member 1 |
| chr18_-_67872891 | 0.06 |
ENST00000454359.1 ENST00000437017.1 |
RTTN |
rotatin |
| chr6_+_116601265 | 0.06 |
ENST00000452085.3 |
DSE |
dermatan sulfate epimerase |
| chr12_-_57522813 | 0.06 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr10_-_128359074 | 0.06 |
ENST00000544758.1 |
C10orf90 |
chromosome 10 open reading frame 90 |
| chr14_-_106330458 | 0.06 |
ENST00000461719.1 |
IGHJ4 |
immunoglobulin heavy joining 4 |
| chr10_-_10836865 | 0.06 |
ENST00000446372.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr18_+_61575200 | 0.06 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr14_+_74077649 | 0.06 |
ENST00000554229.1 |
ACOT6 |
acyl-CoA thioesterase 6 |
| chr7_-_44613494 | 0.05 |
ENST00000431640.1 ENST00000258772.5 |
DDX56 |
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr10_-_10836919 | 0.05 |
ENST00000602763.1 ENST00000415590.2 ENST00000434919.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr7_-_37488834 | 0.05 |
ENST00000310758.4 |
ELMO1 |
engulfment and cell motility 1 |
| chr8_-_141645645 | 0.05 |
ENST00000519980.1 ENST00000220592.5 |
AGO2 |
argonaute RISC catalytic component 2 |
| chr14_+_52734401 | 0.05 |
ENST00000306051.2 ENST00000553372.1 |
PTGDR |
prostaglandin D2 receptor (DP) |
| chr12_-_118112196 | 0.05 |
ENST00000302438.5 |
KSR2 |
kinase suppressor of ras 2 |
| chr16_-_838329 | 0.05 |
ENST00000563560.1 ENST00000569601.1 ENST00000565809.1 ENST00000565377.1 ENST00000007264.2 ENST00000567114.1 |
RPUSD1 |
RNA pseudouridylate synthase domain containing 1 |
| chr1_+_152627927 | 0.05 |
ENST00000444515.1 ENST00000536536.1 |
LINC00302 |
long intergenic non-protein coding RNA 302 |
| chr16_-_28222797 | 0.05 |
ENST00000569951.1 ENST00000565698.1 |
XPO6 |
exportin 6 |
| chrX_-_46759138 | 0.05 |
ENST00000377879.3 |
CXorf31 |
chromosome X open reading frame 31 |
| chr2_-_99870744 | 0.05 |
ENST00000409238.1 ENST00000423800.1 |
LYG2 |
lysozyme G-like 2 |
| chr8_-_95487331 | 0.05 |
ENST00000336148.5 |
RAD54B |
RAD54 homolog B (S. cerevisiae) |
| chr19_+_49661037 | 0.05 |
ENST00000427978.2 |
TRPM4 |
transient receptor potential cation channel, subfamily M, member 4 |
| chr5_+_173472607 | 0.05 |
ENST00000303177.3 ENST00000519867.1 |
NSG2 |
Neuron-specific protein family member 2 |
| chr10_-_29084886 | 0.05 |
ENST00000608061.1 ENST00000443246.2 ENST00000446012.1 |
LINC00837 |
long intergenic non-protein coding RNA 837 |
| chr18_+_72265084 | 0.05 |
ENST00000582337.1 |
ZNF407 |
zinc finger protein 407 |
| chr1_-_234614849 | 0.05 |
ENST00000040877.1 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
| chr20_-_6034672 | 0.05 |
ENST00000378858.4 |
LRRN4 |
leucine rich repeat neuronal 4 |
| chr14_-_82089405 | 0.05 |
ENST00000554211.1 |
RP11-799P8.1 |
RP11-799P8.1 |
| chr2_+_182850551 | 0.05 |
ENST00000452904.1 ENST00000409137.3 ENST00000280295.3 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr4_-_2935674 | 0.05 |
ENST00000514800.1 |
MFSD10 |
major facilitator superfamily domain containing 10 |
| chr11_-_47270341 | 0.05 |
ENST00000529444.1 ENST00000530453.1 ENST00000537863.1 ENST00000529788.1 ENST00000444355.2 ENST00000527256.1 ENST00000529663.1 ENST00000256997.3 |
ACP2 |
acid phosphatase 2, lysosomal |
| chr3_+_50192537 | 0.05 |
ENST00000002829.3 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr16_-_20817753 | 0.05 |
ENST00000389345.5 ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2 |
ERI1 exoribonuclease family member 2 |
| chr17_-_7082861 | 0.05 |
ENST00000269299.3 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr6_-_33756867 | 0.05 |
ENST00000293760.5 |
LEMD2 |
LEM domain containing 2 |
| chr3_+_50192499 | 0.05 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr19_+_38826477 | 0.05 |
ENST00000409410.2 ENST00000215069.4 |
CATSPERG |
catsper channel auxiliary subunit gamma |
| chr12_-_54982300 | 0.05 |
ENST00000547431.1 |
PPP1R1A |
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr19_-_4338838 | 0.04 |
ENST00000594605.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr1_-_120439112 | 0.04 |
ENST00000369400.1 |
ADAM30 |
ADAM metallopeptidase domain 30 |
| chr19_-_4338783 | 0.04 |
ENST00000601482.1 ENST00000600324.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr12_+_93963590 | 0.04 |
ENST00000340600.2 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr19_+_49661079 | 0.04 |
ENST00000355712.5 |
TRPM4 |
transient receptor potential cation channel, subfamily M, member 4 |
| chr19_+_38826415 | 0.04 |
ENST00000410018.1 ENST00000409235.3 |
CATSPERG |
catsper channel auxiliary subunit gamma |
| chr4_+_107236722 | 0.04 |
ENST00000442366.1 |
AIMP1 |
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr14_-_21737610 | 0.04 |
ENST00000320084.7 ENST00000449098.1 ENST00000336053.6 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr11_+_31391381 | 0.04 |
ENST00000465995.1 ENST00000536040.1 |
DNAJC24 |
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr19_+_54496132 | 0.04 |
ENST00000346968.2 |
CACNG6 |
calcium channel, voltage-dependent, gamma subunit 6 |
| chr19_+_49660997 | 0.04 |
ENST00000598691.1 ENST00000252826.5 |
TRPM4 |
transient receptor potential cation channel, subfamily M, member 4 |
| chrX_+_129040094 | 0.04 |
ENST00000425117.2 |
UTP14A |
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr17_-_7082668 | 0.04 |
ENST00000573083.1 ENST00000574388.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr1_+_109656579 | 0.04 |
ENST00000526264.1 ENST00000369939.3 |
KIAA1324 |
KIAA1324 |
| chr15_+_45694523 | 0.04 |
ENST00000305560.6 |
SPATA5L1 |
spermatogenesis associated 5-like 1 |
| chr10_+_6625733 | 0.04 |
ENST00000607982.1 ENST00000608526.1 |
PRKCQ-AS1 |
PRKCQ antisense RNA 1 |
| chr11_+_120107344 | 0.04 |
ENST00000260264.4 |
POU2F3 |
POU class 2 homeobox 3 |
| chr16_+_70557685 | 0.04 |
ENST00000302516.5 ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3 |
splicing factor 3b, subunit 3, 130kDa |
| chr8_-_140715294 | 0.04 |
ENST00000303015.1 ENST00000520439.1 |
KCNK9 |
potassium channel, subfamily K, member 9 |
| chr17_+_26662597 | 0.04 |
ENST00000544907.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr22_+_36044411 | 0.04 |
ENST00000409652.4 |
APOL6 |
apolipoprotein L, 6 |
| chr6_+_106959718 | 0.04 |
ENST00000369066.3 |
AIM1 |
absent in melanoma 1 |
| chr1_+_111770278 | 0.04 |
ENST00000369748.4 |
CHI3L2 |
chitinase 3-like 2 |
| chr3_-_113465065 | 0.04 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr4_+_107236847 | 0.04 |
ENST00000358008.3 |
AIMP1 |
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr2_-_55277692 | 0.04 |
ENST00000394611.2 |
RTN4 |
reticulon 4 |
| chr21_-_36260980 | 0.04 |
ENST00000344691.4 ENST00000358356.5 |
RUNX1 |
runt-related transcription factor 1 |
| chr5_+_34757309 | 0.04 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr11_-_130786400 | 0.04 |
ENST00000265909.4 |
SNX19 |
sorting nexin 19 |
| chr18_-_72264805 | 0.04 |
ENST00000577806.1 |
LINC00909 |
long intergenic non-protein coding RNA 909 |
| chr2_-_55277654 | 0.04 |
ENST00000337526.6 ENST00000317610.7 ENST00000357732.4 |
RTN4 |
reticulon 4 |
| chr1_+_111770232 | 0.04 |
ENST00000369744.2 |
CHI3L2 |
chitinase 3-like 2 |
| chr19_+_3933085 | 0.04 |
ENST00000168977.2 ENST00000599576.1 |
NMRK2 |
nicotinamide riboside kinase 2 |
| chr2_-_10952922 | 0.04 |
ENST00000272227.3 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
| chr18_+_56338750 | 0.03 |
ENST00000345724.3 |
MALT1 |
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr12_+_49687425 | 0.03 |
ENST00000257860.4 |
PRPH |
peripherin |
| chr22_+_38453207 | 0.03 |
ENST00000404072.3 ENST00000424694.1 |
PICK1 |
protein interacting with PRKCA 1 |
| chr2_+_233562015 | 0.03 |
ENST00000427233.1 ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
| chr21_+_34638656 | 0.03 |
ENST00000290200.2 |
IL10RB |
interleukin 10 receptor, beta |
| chr17_+_68071389 | 0.03 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_-_42031300 | 0.03 |
ENST00000592796.1 |
PYY |
peptide YY |
| chr16_-_2059797 | 0.03 |
ENST00000563630.1 |
ZNF598 |
zinc finger protein 598 |
| chr2_-_55277512 | 0.03 |
ENST00000402434.2 |
RTN4 |
reticulon 4 |
| chr22_+_38453378 | 0.03 |
ENST00000437453.1 ENST00000356976.3 |
PICK1 |
protein interacting with PRKCA 1 |
| chr12_+_52345448 | 0.03 |
ENST00000257963.4 ENST00000541224.1 ENST00000426655.2 ENST00000536420.1 ENST00000415850.2 |
ACVR1B |
activin A receptor, type IB |
| chr8_-_54755459 | 0.03 |
ENST00000524234.1 ENST00000521275.1 ENST00000396774.2 |
ATP6V1H |
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr1_-_21503337 | 0.03 |
ENST00000400422.1 ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr9_-_111882195 | 0.03 |
ENST00000374586.3 |
TMEM245 |
transmembrane protein 245 |
| chr17_+_68071458 | 0.03 |
ENST00000589377.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_+_93964158 | 0.03 |
ENST00000549206.1 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr2_-_10952832 | 0.03 |
ENST00000540494.1 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
| chr2_+_33661382 | 0.03 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr15_-_65903574 | 0.03 |
ENST00000420799.2 ENST00000313182.2 ENST00000431261.2 ENST00000442903.3 |
VWA9 |
von Willebrand factor A domain containing 9 |
| chr16_-_28223166 | 0.03 |
ENST00000304658.5 |
XPO6 |
exportin 6 |
| chr19_+_3933579 | 0.03 |
ENST00000593949.1 |
NMRK2 |
nicotinamide riboside kinase 2 |
| chr22_+_23161491 | 0.03 |
ENST00000390316.2 |
IGLV3-9 |
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr14_+_73603126 | 0.03 |
ENST00000557356.1 ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1 |
presenilin 1 |
| chr18_+_56338618 | 0.03 |
ENST00000348428.3 |
MALT1 |
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr11_+_46403303 | 0.03 |
ENST00000407067.1 ENST00000395565.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr6_+_146920116 | 0.03 |
ENST00000367493.3 |
ADGB |
androglobin |
| chr3_-_197476560 | 0.03 |
ENST00000273582.5 |
KIAA0226 |
KIAA0226 |
| chr1_-_85040090 | 0.03 |
ENST00000370630.5 |
CTBS |
chitobiase, di-N-acetyl- |
| chr2_-_55277436 | 0.03 |
ENST00000354474.6 |
RTN4 |
reticulon 4 |
| chr1_-_217311090 | 0.03 |
ENST00000493603.1 ENST00000366940.2 |
ESRRG |
estrogen-related receptor gamma |
| chr11_-_57004658 | 0.03 |
ENST00000606794.1 |
APLNR |
apelin receptor |
| chr9_-_139965000 | 0.03 |
ENST00000409687.3 |
SAPCD2 |
suppressor APC domain containing 2 |
| chr9_+_71736177 | 0.03 |
ENST00000606364.1 ENST00000453658.2 |
TJP2 |
tight junction protein 2 |
| chr18_+_12308231 | 0.03 |
ENST00000590103.1 ENST00000591909.1 ENST00000586653.1 ENST00000592683.1 ENST00000590967.1 ENST00000591208.1 ENST00000591463.1 |
TUBB6 |
tubulin, beta 6 class V |
| chr6_+_146920097 | 0.03 |
ENST00000397944.3 ENST00000522242.1 |
ADGB |
androglobin |
| chr3_+_63263788 | 0.03 |
ENST00000478300.1 |
SYNPR |
synaptoporin |
| chr6_+_35265586 | 0.03 |
ENST00000542066.1 ENST00000316637.5 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
| chr15_+_64752927 | 0.03 |
ENST00000416172.1 |
ZNF609 |
zinc finger protein 609 |
| chr17_+_7211656 | 0.02 |
ENST00000416016.2 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr14_-_23451845 | 0.02 |
ENST00000262713.2 |
AJUBA |
ajuba LIM protein |
| chr11_-_133826852 | 0.02 |
ENST00000533871.2 ENST00000321016.8 |
IGSF9B |
immunoglobulin superfamily, member 9B |
| chr2_-_172290482 | 0.02 |
ENST00000442541.1 ENST00000392599.2 ENST00000375258.4 |
METTL8 |
methyltransferase like 8 |
| chr22_+_32754139 | 0.02 |
ENST00000382088.3 |
RFPL3 |
ret finger protein-like 3 |
| chr10_-_5046042 | 0.02 |
ENST00000421196.3 ENST00000455190.1 |
AKR1C2 |
aldo-keto reductase family 1, member C2 |
| chr19_-_5839703 | 0.02 |
ENST00000524754.1 |
FUT6 |
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr19_-_5839721 | 0.02 |
ENST00000286955.5 ENST00000318336.4 |
FUT6 |
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr17_+_68100989 | 0.02 |
ENST00000585558.1 ENST00000392670.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr16_-_2059748 | 0.02 |
ENST00000562103.1 ENST00000431526.1 |
ZNF598 |
zinc finger protein 598 |
| chr11_+_46403194 | 0.02 |
ENST00000395569.4 ENST00000395566.4 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr6_+_157099036 | 0.02 |
ENST00000350026.5 ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
| chr11_+_46402744 | 0.02 |
ENST00000533952.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.2 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.3 | GO:2000391 | positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.1 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.0 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.2 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.0 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |