Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
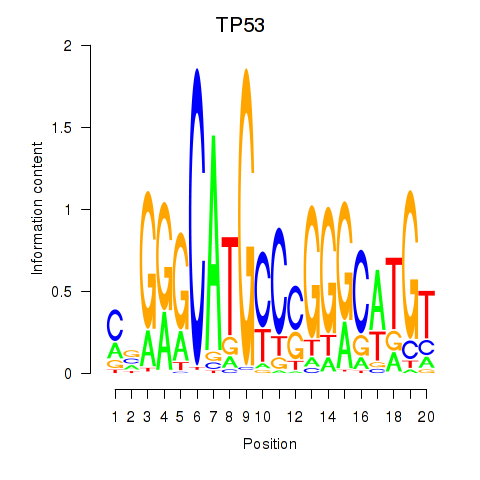
Results for TP53
Z-value: 1.56
Transcription factors associated with TP53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP53
|
ENSG00000141510.11 | TP53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP53 | hg19_v2_chr17_-_7590745_7590856 | 0.24 | 5.7e-01 | Click! |
Activity profile of TP53 motif
Sorted Z-values of TP53 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TP53
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51504411 | 2.00 |
ENST00000593490.1 |
KLK8 |
kallikrein-related peptidase 8 |
| chr1_-_153588334 | 1.94 |
ENST00000476873.1 |
S100A14 |
S100 calcium binding protein A14 |
| chr18_-_28622774 | 1.65 |
ENST00000434452.1 |
DSC3 |
desmocollin 3 |
| chr18_-_28622699 | 1.64 |
ENST00000360428.4 |
DSC3 |
desmocollin 3 |
| chr6_+_36098262 | 1.49 |
ENST00000373761.6 ENST00000373766.5 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chr10_+_48255253 | 1.35 |
ENST00000357718.4 ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8 AL591684.1 |
annexin A8 Protein LOC100996760 |
| chr15_+_40532058 | 1.31 |
ENST00000260404.4 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr10_+_47746929 | 1.29 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr8_-_10588010 | 1.16 |
ENST00000304501.1 |
SOX7 |
SRY (sex determining region Y)-box 7 |
| chr3_-_12800751 | 0.97 |
ENST00000435218.2 ENST00000435575.1 |
TMEM40 |
transmembrane protein 40 |
| chr11_-_108464465 | 0.97 |
ENST00000525344.1 |
EXPH5 |
exophilin 5 |
| chr1_-_209825674 | 0.94 |
ENST00000367030.3 ENST00000356082.4 |
LAMB3 |
laminin, beta 3 |
| chr11_-_108464321 | 0.93 |
ENST00000265843.4 |
EXPH5 |
exophilin 5 |
| chr6_+_36097992 | 0.83 |
ENST00000211287.4 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chr10_-_47173994 | 0.78 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr16_+_57653989 | 0.78 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr16_+_57653854 | 0.77 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr13_+_37006421 | 0.71 |
ENST00000255465.4 |
CCNA1 |
cyclin A1 |
| chr13_+_37006398 | 0.70 |
ENST00000418263.1 |
CCNA1 |
cyclin A1 |
| chr9_-_135996537 | 0.68 |
ENST00000372050.3 ENST00000372047.3 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
| chr6_-_30712313 | 0.68 |
ENST00000376377.2 ENST00000259874.5 |
IER3 |
immediate early response 3 |
| chr6_-_138428613 | 0.67 |
ENST00000421351.3 |
PERP |
PERP, TP53 apoptosis effector |
| chr8_-_23082580 | 0.65 |
ENST00000221132.3 |
TNFRSF10A |
tumor necrosis factor receptor superfamily, member 10a |
| chr11_+_1856034 | 0.63 |
ENST00000341958.3 |
SYT8 |
synaptotagmin VIII |
| chr12_+_44229846 | 0.60 |
ENST00000551577.1 ENST00000266534.3 |
TMEM117 |
transmembrane protein 117 |
| chr12_-_126467906 | 0.58 |
ENST00000507313.1 ENST00000545784.1 |
LINC00939 |
long intergenic non-protein coding RNA 939 |
| chr16_+_57406368 | 0.58 |
ENST00000006053.6 ENST00000563383.1 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
| chr1_+_13910479 | 0.56 |
ENST00000509009.1 |
PDPN |
podoplanin |
| chr11_+_706595 | 0.55 |
ENST00000531348.1 ENST00000530636.1 |
EPS8L2 |
EPS8-like 2 |
| chr11_+_1855645 | 0.51 |
ENST00000381968.3 ENST00000381978.3 |
SYT8 |
synaptotagmin VIII |
| chr11_-_44972476 | 0.51 |
ENST00000527685.1 ENST00000308212.5 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr1_+_13910757 | 0.50 |
ENST00000376061.4 ENST00000513143.1 |
PDPN |
podoplanin |
| chr11_-_44972390 | 0.49 |
ENST00000395648.3 ENST00000531928.2 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chrX_+_37545012 | 0.49 |
ENST00000378616.3 |
XK |
X-linked Kx blood group (McLeod syndrome) |
| chr19_+_3136115 | 0.47 |
ENST00000262958.3 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
| chr1_-_201438282 | 0.45 |
ENST00000367311.3 ENST00000367309.1 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
| chr4_-_48908737 | 0.44 |
ENST00000381464.2 |
OCIAD2 |
OCIA domain containing 2 |
| chr1_+_13910194 | 0.43 |
ENST00000376057.4 ENST00000510906.1 |
PDPN |
podoplanin |
| chr4_+_103422471 | 0.41 |
ENST00000226574.4 ENST00000394820.4 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr8_-_22926526 | 0.39 |
ENST00000347739.3 ENST00000542226.1 |
TNFRSF10B |
tumor necrosis factor receptor superfamily, member 10b |
| chr1_-_38412683 | 0.39 |
ENST00000373024.3 ENST00000373023.2 |
INPP5B |
inositol polyphosphate-5-phosphatase, 75kDa |
| chr4_-_48908805 | 0.38 |
ENST00000273860.4 |
OCIAD2 |
OCIA domain containing 2 |
| chr1_-_10532531 | 0.38 |
ENST00000377036.2 ENST00000377038.3 |
DFFA |
DNA fragmentation factor, 45kDa, alpha polypeptide |
| chr4_-_48908822 | 0.37 |
ENST00000508632.1 |
OCIAD2 |
OCIA domain containing 2 |
| chr22_+_38302285 | 0.36 |
ENST00000215957.6 |
MICALL1 |
MICAL-like 1 |
| chr11_-_59383617 | 0.34 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr2_-_75788038 | 0.33 |
ENST00000393913.3 ENST00000410113.1 |
EVA1A |
eva-1 homolog A (C. elegans) |
| chr7_-_72439997 | 0.33 |
ENST00000285805.3 |
TRIM74 |
tripartite motif containing 74 |
| chr8_-_22926623 | 0.33 |
ENST00000276431.4 |
TNFRSF10B |
tumor necrosis factor receptor superfamily, member 10b |
| chr2_+_233734994 | 0.32 |
ENST00000331342.2 |
C2orf82 |
chromosome 2 open reading frame 82 |
| chr9_+_139847347 | 0.32 |
ENST00000371632.3 |
LCN12 |
lipocalin 12 |
| chr3_+_184279566 | 0.31 |
ENST00000330394.2 |
EPHB3 |
EPH receptor B3 |
| chr2_-_26205340 | 0.30 |
ENST00000264712.3 |
KIF3C |
kinesin family member 3C |
| chr9_+_138606400 | 0.30 |
ENST00000486577.2 |
KCNT1 |
potassium channel, subfamily T, member 1 |
| chr1_+_11796126 | 0.29 |
ENST00000376637.3 |
AGTRAP |
angiotensin II receptor-associated protein |
| chr1_+_11796177 | 0.28 |
ENST00000400895.2 ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP |
angiotensin II receptor-associated protein |
| chr4_-_153457197 | 0.28 |
ENST00000281708.4 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr17_-_74528128 | 0.28 |
ENST00000590175.1 |
CYGB |
cytoglobin |
| chr5_-_95297534 | 0.27 |
ENST00000513343.1 ENST00000431061.2 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr6_+_116692102 | 0.26 |
ENST00000359564.2 |
DSE |
dermatan sulfate epimerase |
| chr22_-_18257178 | 0.26 |
ENST00000342111.5 |
BID |
BH3 interacting domain death agonist |
| chrX_+_99899180 | 0.26 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr1_-_156051789 | 0.25 |
ENST00000532414.2 |
MEX3A |
mex-3 RNA binding family member A |
| chr11_+_64059464 | 0.24 |
ENST00000394525.2 |
KCNK4 |
potassium channel, subfamily K, member 4 |
| chr3_+_122785895 | 0.24 |
ENST00000316218.7 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
| chr3_-_184079382 | 0.24 |
ENST00000344937.7 ENST00000423355.2 ENST00000434054.2 ENST00000457512.1 ENST00000265593.4 |
CLCN2 |
chloride channel, voltage-sensitive 2 |
| chr1_+_10057274 | 0.24 |
ENST00000294435.7 |
RBP7 |
retinol binding protein 7, cellular |
| chr1_+_116519112 | 0.24 |
ENST00000369503.4 |
SLC22A15 |
solute carrier family 22, member 15 |
| chr6_+_4776580 | 0.23 |
ENST00000397588.3 |
CDYL |
chromodomain protein, Y-like |
| chr14_+_97263641 | 0.23 |
ENST00000216639.3 |
VRK1 |
vaccinia related kinase 1 |
| chr19_-_51530916 | 0.23 |
ENST00000594768.1 |
KLK11 |
kallikrein-related peptidase 11 |
| chr20_+_42984330 | 0.23 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr19_-_42746714 | 0.23 |
ENST00000222330.3 |
GSK3A |
glycogen synthase kinase 3 alpha |
| chr9_-_138853156 | 0.22 |
ENST00000371756.3 |
UBAC1 |
UBA domain containing 1 |
| chr6_+_24403144 | 0.21 |
ENST00000274747.7 ENST00000543597.1 ENST00000535061.1 ENST00000378353.1 ENST00000378386.3 ENST00000443868.2 |
MRS2 |
MRS2 magnesium transporter |
| chr11_+_100558384 | 0.21 |
ENST00000524892.2 ENST00000298815.8 |
ARHGAP42 |
Rho GTPase activating protein 42 |
| chr6_-_101329157 | 0.21 |
ENST00000369143.2 |
ASCC3 |
activating signal cointegrator 1 complex subunit 3 |
| chr11_+_64126614 | 0.21 |
ENST00000528057.1 ENST00000334205.4 ENST00000294261.4 |
RPS6KA4 |
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr19_-_47735918 | 0.20 |
ENST00000449228.1 ENST00000300880.7 ENST00000341983.4 |
BBC3 |
BCL2 binding component 3 |
| chr12_-_64784471 | 0.20 |
ENST00000333722.5 |
C12orf56 |
chromosome 12 open reading frame 56 |
| chr1_-_1356719 | 0.19 |
ENST00000520296.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr17_+_74372662 | 0.19 |
ENST00000591651.1 ENST00000545180.1 |
SPHK1 |
sphingosine kinase 1 |
| chr19_+_39687596 | 0.18 |
ENST00000339852.4 |
NCCRP1 |
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr1_-_205744574 | 0.18 |
ENST00000367139.3 ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr4_+_41937131 | 0.17 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr3_+_184079492 | 0.17 |
ENST00000456318.1 ENST00000412877.1 ENST00000438240.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr4_-_187644930 | 0.17 |
ENST00000441802.2 |
FAT1 |
FAT atypical cadherin 1 |
| chr19_+_56652556 | 0.17 |
ENST00000337080.3 |
ZNF444 |
zinc finger protein 444 |
| chr2_+_169312350 | 0.17 |
ENST00000305747.6 |
CERS6 |
ceramide synthase 6 |
| chr10_-_105212059 | 0.17 |
ENST00000260743.5 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr1_-_205744205 | 0.16 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr22_+_29279552 | 0.16 |
ENST00000544604.2 |
ZNRF3 |
zinc and ring finger 3 |
| chr2_+_169312725 | 0.16 |
ENST00000392687.4 |
CERS6 |
ceramide synthase 6 |
| chr11_-_3078616 | 0.15 |
ENST00000401769.3 ENST00000278224.9 ENST00000397114.3 ENST00000380525.4 |
CARS |
cysteinyl-tRNA synthetase |
| chr13_-_48877795 | 0.15 |
ENST00000436963.1 ENST00000433480.2 |
LINC00441 |
long intergenic non-protein coding RNA 441 |
| chr1_-_1356628 | 0.15 |
ENST00000442470.1 ENST00000537107.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr19_-_17014408 | 0.14 |
ENST00000594249.1 |
CPAMD8 |
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr12_+_124086632 | 0.14 |
ENST00000238146.4 ENST00000538744.1 |
DDX55 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr8_-_109799793 | 0.14 |
ENST00000297459.3 |
TMEM74 |
transmembrane protein 74 |
| chr1_+_25598989 | 0.14 |
ENST00000454452.2 |
RHD |
Rh blood group, D antigen |
| chr10_-_105212141 | 0.14 |
ENST00000369788.3 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr11_-_44972418 | 0.13 |
ENST00000525680.1 ENST00000528290.1 ENST00000530035.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr7_-_56101826 | 0.13 |
ENST00000421626.1 |
PSPH |
phosphoserine phosphatase |
| chr20_-_1373606 | 0.13 |
ENST00000381715.1 ENST00000439640.2 ENST00000381719.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr11_+_36397528 | 0.13 |
ENST00000311599.5 ENST00000378867.3 |
PRR5L |
proline rich 5 like |
| chr12_+_12966250 | 0.13 |
ENST00000352940.4 ENST00000358007.3 ENST00000544400.1 |
DDX47 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr3_-_46037299 | 0.13 |
ENST00000296137.2 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
| chr1_+_211432700 | 0.13 |
ENST00000452621.2 |
RCOR3 |
REST corepressor 3 |
| chr1_-_25747283 | 0.12 |
ENST00000346452.4 ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE |
Rh blood group, CcEe antigens |
| chr11_-_3078838 | 0.12 |
ENST00000397111.5 |
CARS |
cysteinyl-tRNA synthetase |
| chr11_+_119039414 | 0.12 |
ENST00000409991.1 ENST00000292199.2 ENST00000409265.4 ENST00000409109.1 |
NLRX1 |
NLR family member X1 |
| chr17_-_61523622 | 0.12 |
ENST00000448884.2 ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561 |
cytochrome b561 |
| chr14_-_36990354 | 0.12 |
ENST00000518149.1 |
NKX2-1 |
NK2 homeobox 1 |
| chr5_+_72861560 | 0.12 |
ENST00000296792.4 ENST00000509005.1 ENST00000543251.1 ENST00000508686.1 ENST00000508491.1 |
UTP15 |
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
| chr19_+_56652686 | 0.12 |
ENST00000592949.1 |
ZNF444 |
zinc finger protein 444 |
| chr3_+_23851928 | 0.12 |
ENST00000467766.1 ENST00000424381.1 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr4_-_159644507 | 0.11 |
ENST00000307720.3 |
PPID |
peptidylprolyl isomerase D |
| chr22_-_18257249 | 0.11 |
ENST00000399765.1 ENST00000399767.1 ENST00000399774.3 |
BID |
BH3 interacting domain death agonist |
| chr2_+_95691417 | 0.11 |
ENST00000309988.4 |
MAL |
mal, T-cell differentiation protein |
| chr19_+_13229126 | 0.11 |
ENST00000292431.4 |
NACC1 |
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr9_-_95527079 | 0.11 |
ENST00000356884.6 ENST00000375512.3 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
| chr5_+_121647877 | 0.11 |
ENST00000514497.2 ENST00000261367.7 |
SNCAIP |
synuclein, alpha interacting protein |
| chr11_+_33278811 | 0.11 |
ENST00000303296.4 ENST00000379016.3 |
HIPK3 |
homeodomain interacting protein kinase 3 |
| chr11_-_47400062 | 0.11 |
ENST00000533030.1 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
| chrX_-_48827976 | 0.11 |
ENST00000218176.3 |
KCND1 |
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr17_-_61523535 | 0.11 |
ENST00000584031.1 ENST00000392976.1 |
CYB561 |
cytochrome b561 |
| chr2_+_95691445 | 0.11 |
ENST00000353004.3 ENST00000354078.3 ENST00000349807.3 |
MAL |
mal, T-cell differentiation protein |
| chr19_-_42721819 | 0.11 |
ENST00000336034.4 ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2 |
death effector domain containing 2 |
| chr22_+_29168652 | 0.11 |
ENST00000249064.4 ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117 |
coiled-coil domain containing 117 |
| chr12_+_53894672 | 0.11 |
ENST00000266987.2 ENST00000456234.2 |
TARBP2 |
TAR (HIV-1) RNA binding protein 2 |
| chr5_+_121647764 | 0.11 |
ENST00000261368.8 ENST00000379533.2 ENST00000379536.2 ENST00000379538.3 |
SNCAIP |
synuclein, alpha interacting protein |
| chr19_-_6481776 | 0.11 |
ENST00000543576.1 ENST00000590173.1 ENST00000381480.2 |
DENND1C |
DENN/MADD domain containing 1C |
| chr11_-_77122928 | 0.10 |
ENST00000528203.1 ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr1_-_26197744 | 0.10 |
ENST00000374296.3 |
PAQR7 |
progestin and adipoQ receptor family member VII |
| chr11_+_61717279 | 0.10 |
ENST00000378043.4 |
BEST1 |
bestrophin 1 |
| chr19_+_42746927 | 0.10 |
ENST00000378108.1 |
AC006486.1 |
AC006486.1 |
| chr19_-_55690758 | 0.10 |
ENST00000590851.1 |
SYT5 |
synaptotagmin V |
| chr1_+_26485511 | 0.10 |
ENST00000374268.3 |
FAM110D |
family with sequence similarity 110, member D |
| chr14_-_64971288 | 0.10 |
ENST00000394715.1 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
| chr1_+_42846443 | 0.10 |
ENST00000410070.2 ENST00000431473.3 |
RIMKLA |
ribosomal modification protein rimK-like family member A |
| chr12_+_53895364 | 0.10 |
ENST00000552817.1 ENST00000394357.2 |
TARBP2 |
TAR (HIV-1) RNA binding protein 2 |
| chr1_+_243419306 | 0.10 |
ENST00000355875.4 ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8 |
serologically defined colon cancer antigen 8 |
| chr19_-_52150053 | 0.10 |
ENST00000599649.1 ENST00000429354.3 ENST00000360844.6 ENST00000222107.4 |
SIGLEC5 SIGLEC14 SIGLEC5 |
SIGLEC5 sialic acid binding Ig-like lectin 14 sialic acid binding Ig-like lectin 5 |
| chr17_+_18087553 | 0.10 |
ENST00000399138.4 |
ALKBH5 |
alkB, alkylation repair homolog 5 (E. coli) |
| chr13_+_31480328 | 0.10 |
ENST00000380482.4 |
MEDAG |
mesenteric estrogen-dependent adipogenesis |
| chr14_-_64970494 | 0.10 |
ENST00000608382.1 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
| chr12_+_53895052 | 0.10 |
ENST00000552857.1 |
TARBP2 |
TAR (HIV-1) RNA binding protein 2 |
| chr2_+_47168630 | 0.09 |
ENST00000263737.6 |
TTC7A |
tetratricopeptide repeat domain 7A |
| chr1_-_202936394 | 0.09 |
ENST00000367249.4 |
CYB5R1 |
cytochrome b5 reductase 1 |
| chr1_-_150738261 | 0.09 |
ENST00000448301.2 ENST00000368985.3 |
CTSS |
cathepsin S |
| chr3_-_113465065 | 0.09 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr13_-_20735178 | 0.09 |
ENST00000241125.3 |
GJA3 |
gap junction protein, alpha 3, 46kDa |
| chr16_-_4303767 | 0.09 |
ENST00000573268.1 ENST00000573042.1 |
RP11-95P2.1 |
RP11-95P2.1 |
| chr11_+_61717336 | 0.09 |
ENST00000378042.3 |
BEST1 |
bestrophin 1 |
| chr2_+_85661918 | 0.09 |
ENST00000340326.2 |
SH2D6 |
SH2 domain containing 6 |
| chr5_-_72861484 | 0.09 |
ENST00000296785.3 |
ANKRA2 |
ankyrin repeat, family A (RFXANK-like), 2 |
| chr8_-_124253576 | 0.09 |
ENST00000276704.4 |
C8orf76 |
chromosome 8 open reading frame 76 |
| chr6_-_114292449 | 0.09 |
ENST00000519065.1 |
HDAC2 |
histone deacetylase 2 |
| chr11_-_129062093 | 0.08 |
ENST00000310343.9 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr2_-_242556900 | 0.08 |
ENST00000402545.1 ENST00000402136.1 |
THAP4 |
THAP domain containing 4 |
| chr9_-_104160872 | 0.08 |
ENST00000539624.1 ENST00000374865.4 |
MRPL50 |
mitochondrial ribosomal protein L50 |
| chr5_+_121647386 | 0.08 |
ENST00000542191.1 ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP |
synuclein, alpha interacting protein |
| chr3_+_196466710 | 0.08 |
ENST00000327134.3 |
PAK2 |
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr19_+_2785458 | 0.08 |
ENST00000307741.6 ENST00000585338.1 |
THOP1 |
thimet oligopeptidase 1 |
| chr17_+_18128896 | 0.08 |
ENST00000316843.4 |
LLGL1 |
lethal giant larvae homolog 1 (Drosophila) |
| chr1_-_2458026 | 0.08 |
ENST00000435556.3 ENST00000378466.3 |
PANK4 |
pantothenate kinase 4 |
| chr12_-_113772835 | 0.08 |
ENST00000552014.1 ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr6_-_114292284 | 0.08 |
ENST00000520895.1 ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2 |
histone deacetylase 2 |
| chr11_+_64073699 | 0.08 |
ENST00000405666.1 ENST00000468670.1 |
ESRRA |
estrogen-related receptor alpha |
| chr19_-_1605424 | 0.08 |
ENST00000589880.1 ENST00000585671.1 ENST00000591899.3 |
UQCR11 |
ubiquinol-cytochrome c reductase, complex III subunit XI |
| chr10_-_126847276 | 0.08 |
ENST00000531469.1 |
CTBP2 |
C-terminal binding protein 2 |
| chr3_-_100120223 | 0.08 |
ENST00000284320.5 |
TOMM70A |
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr20_-_23618582 | 0.07 |
ENST00000398411.1 ENST00000376925.3 |
CST3 |
cystatin C |
| chr14_-_102553371 | 0.07 |
ENST00000553585.1 ENST00000216281.8 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr19_-_5567842 | 0.07 |
ENST00000587632.1 |
TINCR |
tissue differentiation-inducing non-protein coding RNA |
| chr6_-_101329191 | 0.07 |
ENST00000324723.6 ENST00000369162.2 ENST00000522650.1 |
ASCC3 |
activating signal cointegrator 1 complex subunit 3 |
| chr1_+_27153173 | 0.07 |
ENST00000374142.4 |
ZDHHC18 |
zinc finger, DHHC-type containing 18 |
| chr9_-_116037840 | 0.06 |
ENST00000374206.3 |
CDC26 |
cell division cycle 26 |
| chr3_-_52479043 | 0.06 |
ENST00000231721.2 ENST00000475739.1 |
SEMA3G |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chrX_-_153285395 | 0.06 |
ENST00000369980.3 |
IRAK1 |
interleukin-1 receptor-associated kinase 1 |
| chr20_-_61002584 | 0.06 |
ENST00000252998.1 |
RBBP8NL |
RBBP8 N-terminal like |
| chr8_-_12051576 | 0.06 |
ENST00000524571.2 ENST00000533852.2 ENST00000533513.1 ENST00000448228.2 ENST00000534520.1 ENST00000321602.8 |
FAM86B1 |
family with sequence similarity 86, member B1 |
| chr5_-_131892501 | 0.06 |
ENST00000450655.1 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr22_+_19705928 | 0.06 |
ENST00000383045.3 ENST00000438754.2 |
SEPT5 |
septin 5 |
| chr11_-_77123065 | 0.06 |
ENST00000530617.1 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr19_+_827823 | 0.06 |
ENST00000233997.2 |
AZU1 |
azurocidin 1 |
| chr19_+_51293672 | 0.06 |
ENST00000270593.1 ENST00000270594.3 |
ACPT |
acid phosphatase, testicular |
| chr1_-_169396666 | 0.06 |
ENST00000456107.1 ENST00000367805.3 |
CCDC181 |
coiled-coil domain containing 181 |
| chr16_-_58231782 | 0.06 |
ENST00000565188.1 ENST00000262506.3 |
CSNK2A2 |
casein kinase 2, alpha prime polypeptide |
| chr2_+_47168313 | 0.06 |
ENST00000319190.5 ENST00000394850.2 ENST00000536057.1 |
TTC7A |
tetratricopeptide repeat domain 7A |
| chr17_-_14140166 | 0.06 |
ENST00000420162.2 ENST00000431716.2 |
CDRT15 |
CMT1A duplicated region transcript 15 |
| chr1_+_54411715 | 0.06 |
ENST00000371370.3 ENST00000371368.1 |
LRRC42 |
leucine rich repeat containing 42 |
| chr19_+_13988061 | 0.05 |
ENST00000339133.5 ENST00000397555.2 |
NANOS3 |
nanos homolog 3 (Drosophila) |
| chr17_-_5095126 | 0.05 |
ENST00000576772.1 ENST00000575779.1 |
ZNF594 |
zinc finger protein 594 |
| chr1_+_25598872 | 0.05 |
ENST00000328664.4 |
RHD |
Rh blood group, D antigen |
| chr11_+_60699222 | 0.05 |
ENST00000536409.1 |
TMEM132A |
transmembrane protein 132A |
| chr1_+_44115814 | 0.05 |
ENST00000372396.3 |
KDM4A |
lysine (K)-specific demethylase 4A |
| chr3_+_149530469 | 0.05 |
ENST00000392894.3 ENST00000344229.3 |
RNF13 |
ring finger protein 13 |
| chr5_+_121647924 | 0.05 |
ENST00000414317.2 |
SNCAIP |
synuclein, alpha interacting protein |
| chr12_-_14721283 | 0.05 |
ENST00000240617.5 |
PLBD1 |
phospholipase B domain containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.3 | 1.3 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.2 | 2.0 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 0.6 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.2 | 0.5 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.4 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.1 | 0.4 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.3 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.1 | 1.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.4 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.1 | 0.4 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 | 0.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.2 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.5 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.6 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 1.9 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.7 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.3 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 1.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.4 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.3 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 3.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.2 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 2.2 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 1.2 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:0045348 | positive regulation of chemokine biosynthetic process(GO:0045080) positive regulation of MHC class II biosynthetic process(GO:0045348) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0060313 | negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 1.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.3 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 1.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 4.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 1.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.3 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 2.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 1.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 2.5 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 2.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0038023 | steroid hormone receptor activity(GO:0003707) signaling receptor activity(GO:0038023) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |