Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for TP63
Z-value: 1.68
Transcription factors associated with TP63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP63
|
ENSG00000073282.8 | TP63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP63 | hg19_v2_chr3_+_189507432_189507459 | 0.89 | 3.2e-03 | Click! |
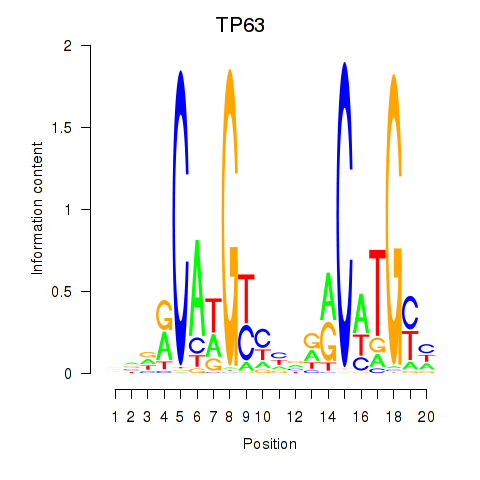
Activity profile of TP63 motif
Sorted Z-values of TP63 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TP63
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51504411 | 3.39 |
ENST00000593490.1 |
KLK8 |
kallikrein-related peptidase 8 |
| chr1_-_153588334 | 2.94 |
ENST00000476873.1 |
S100A14 |
S100 calcium binding protein A14 |
| chr12_-_52845910 | 2.89 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr1_+_86889769 | 2.66 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr12_-_52887034 | 2.62 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr11_+_18287721 | 2.12 |
ENST00000356524.4 |
SAA1 |
serum amyloid A1 |
| chr11_+_18287801 | 2.11 |
ENST00000532858.1 ENST00000405158.2 |
SAA1 |
serum amyloid A1 |
| chr4_-_10023095 | 2.00 |
ENST00000264784.3 |
SLC2A9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr16_+_57653989 | 1.91 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr16_+_57653854 | 1.79 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr1_+_35220613 | 1.70 |
ENST00000338513.1 |
GJB5 |
gap junction protein, beta 5, 31.1kDa |
| chr2_-_216003127 | 1.65 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr3_-_57113314 | 1.64 |
ENST00000338458.4 ENST00000468727.1 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr1_-_209824643 | 1.59 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr15_+_41136216 | 1.56 |
ENST00000562057.1 ENST00000344051.4 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr3_+_122044084 | 1.49 |
ENST00000264474.3 ENST00000479204.1 |
CSTA |
cystatin A (stefin A) |
| chr18_+_34124507 | 1.29 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr15_+_40532058 | 1.14 |
ENST00000260404.4 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr6_+_54711533 | 1.10 |
ENST00000306858.7 |
FAM83B |
family with sequence similarity 83, member B |
| chr9_+_93589734 | 1.09 |
ENST00000375746.1 |
SYK |
spleen tyrosine kinase |
| chr1_-_32801825 | 0.90 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr2_-_31361543 | 0.84 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_+_153747746 | 0.80 |
ENST00000368661.3 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr12_-_126467906 | 0.77 |
ENST00000507313.1 ENST00000545784.1 |
LINC00939 |
long intergenic non-protein coding RNA 939 |
| chr6_+_106546808 | 0.76 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr20_-_6103666 | 0.74 |
ENST00000536936.1 |
FERMT1 |
fermitin family member 1 |
| chr19_-_11688447 | 0.73 |
ENST00000590420.1 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr6_-_30712313 | 0.68 |
ENST00000376377.2 ENST00000259874.5 |
IER3 |
immediate early response 3 |
| chr1_+_24646002 | 0.66 |
ENST00000356046.2 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_24645807 | 0.65 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_24645865 | 0.65 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr22_+_38302285 | 0.63 |
ENST00000215957.6 |
MICALL1 |
MICAL-like 1 |
| chr6_+_35265586 | 0.62 |
ENST00000542066.1 ENST00000316637.5 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
| chr9_+_138606400 | 0.60 |
ENST00000486577.2 |
KCNT1 |
potassium channel, subfamily T, member 1 |
| chr3_-_191000172 | 0.60 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr16_+_82090028 | 0.60 |
ENST00000568090.1 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr3_+_136676851 | 0.57 |
ENST00000309741.5 |
IL20RB |
interleukin 20 receptor beta |
| chr2_-_165698662 | 0.57 |
ENST00000194871.6 ENST00000445474.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr11_+_1856034 | 0.57 |
ENST00000341958.3 |
SYT8 |
synaptotagmin VIII |
| chr3_+_136676707 | 0.56 |
ENST00000329582.4 |
IL20RB |
interleukin 20 receptor beta |
| chr2_-_165698521 | 0.55 |
ENST00000409184.3 ENST00000392717.2 ENST00000456693.1 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr12_-_15114492 | 0.55 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr6_+_121756809 | 0.54 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr17_-_34079897 | 0.54 |
ENST00000254466.6 ENST00000587565.1 |
GAS2L2 |
growth arrest-specific 2 like 2 |
| chr12_-_15114603 | 0.53 |
ENST00000228945.4 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr18_-_28742813 | 0.50 |
ENST00000257197.3 ENST00000257198.5 |
DSC1 |
desmocollin 1 |
| chr11_-_104972158 | 0.49 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr17_+_1646130 | 0.48 |
ENST00000453066.1 ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr12_-_15114658 | 0.48 |
ENST00000542276.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr9_+_71944241 | 0.46 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr1_-_1356719 | 0.45 |
ENST00000520296.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr19_-_11688500 | 0.43 |
ENST00000433365.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr8_-_22926526 | 0.42 |
ENST00000347739.3 ENST00000542226.1 |
TNFRSF10B |
tumor necrosis factor receptor superfamily, member 10b |
| chr1_+_42619070 | 0.41 |
ENST00000372581.1 |
GUCA2B |
guanylate cyclase activator 2B (uroguanylin) |
| chr2_+_11752379 | 0.41 |
ENST00000396123.1 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr9_+_101569944 | 0.40 |
ENST00000375011.3 |
GALNT12 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) |
| chr19_-_19051103 | 0.40 |
ENST00000542541.2 ENST00000433218.2 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr8_+_74206829 | 0.39 |
ENST00000240285.5 |
RDH10 |
retinol dehydrogenase 10 (all-trans) |
| chr1_-_1356628 | 0.39 |
ENST00000442470.1 ENST00000537107.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr17_+_26662597 | 0.38 |
ENST00000544907.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr4_+_95917383 | 0.38 |
ENST00000512312.1 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr20_-_43280361 | 0.38 |
ENST00000372874.4 |
ADA |
adenosine deaminase |
| chr7_-_72439997 | 0.37 |
ENST00000285805.3 |
TRIM74 |
tripartite motif containing 74 |
| chr22_+_50628999 | 0.36 |
ENST00000395827.1 |
TRABD |
TraB domain containing |
| chr1_-_152552980 | 0.36 |
ENST00000368787.3 |
LCE3D |
late cornified envelope 3D |
| chr1_-_118727781 | 0.36 |
ENST00000336338.5 |
SPAG17 |
sperm associated antigen 17 |
| chr8_-_22926623 | 0.36 |
ENST00000276431.4 |
TNFRSF10B |
tumor necrosis factor receptor superfamily, member 10b |
| chr19_-_51530916 | 0.34 |
ENST00000594768.1 |
KLK11 |
kallikrein-related peptidase 11 |
| chr6_-_101329157 | 0.34 |
ENST00000369143.2 |
ASCC3 |
activating signal cointegrator 1 complex subunit 3 |
| chr16_+_19467772 | 0.34 |
ENST00000219821.5 ENST00000561503.1 ENST00000564959.1 |
TMC5 |
transmembrane channel-like 5 |
| chr6_+_18387570 | 0.33 |
ENST00000259939.3 |
RNF144B |
ring finger protein 144B |
| chr11_-_104905840 | 0.33 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr20_-_43743790 | 0.32 |
ENST00000307971.4 ENST00000372789.4 |
WFDC5 |
WAP four-disulfide core domain 5 |
| chr1_-_26197744 | 0.32 |
ENST00000374296.3 |
PAQR7 |
progestin and adipoQ receptor family member VII |
| chrX_-_8700171 | 0.32 |
ENST00000262648.3 |
KAL1 |
Kallmann syndrome 1 sequence |
| chr3_+_184080387 | 0.32 |
ENST00000455712.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr1_+_17575584 | 0.31 |
ENST00000375460.3 |
PADI3 |
peptidyl arginine deiminase, type III |
| chr11_-_72433346 | 0.31 |
ENST00000334211.8 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr3_-_33700589 | 0.31 |
ENST00000461133.3 ENST00000496954.2 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr1_+_26485511 | 0.31 |
ENST00000374268.3 |
FAM110D |
family with sequence similarity 110, member D |
| chr20_-_43280325 | 0.30 |
ENST00000537820.1 |
ADA |
adenosine deaminase |
| chr11_+_19138670 | 0.30 |
ENST00000446113.2 ENST00000399351.3 |
ZDHHC13 |
zinc finger, DHHC-type containing 13 |
| chr19_+_56459198 | 0.30 |
ENST00000291971.3 ENST00000590542.1 |
NLRP8 |
NLR family, pyrin domain containing 8 |
| chr19_-_51471381 | 0.30 |
ENST00000594641.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr12_+_110172572 | 0.29 |
ENST00000358906.3 |
FAM222A |
family with sequence similarity 222, member A |
| chr10_+_99344071 | 0.29 |
ENST00000370647.4 ENST00000370646.4 |
HOGA1 |
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr19_-_47734448 | 0.28 |
ENST00000439096.2 |
BBC3 |
BCL2 binding component 3 |
| chr19_-_11639931 | 0.28 |
ENST00000592312.1 ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT |
ECSIT signalling integrator |
| chr6_+_134758827 | 0.27 |
ENST00000431422.1 |
LINC01010 |
long intergenic non-protein coding RNA 1010 |
| chr19_+_15160130 | 0.27 |
ENST00000427043.3 |
CASP14 |
caspase 14, apoptosis-related cysteine peptidase |
| chr11_-_72432950 | 0.27 |
ENST00000426523.1 ENST00000429686.1 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr3_+_184081213 | 0.26 |
ENST00000429568.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr19_+_18496957 | 0.26 |
ENST00000252809.3 |
GDF15 |
growth differentiation factor 15 |
| chr11_-_77850629 | 0.26 |
ENST00000376156.3 ENST00000525870.1 ENST00000530454.1 ENST00000525755.1 ENST00000527099.1 ENST00000525761.1 ENST00000299626.5 |
ALG8 |
ALG8, alpha-1,3-glucosyltransferase |
| chr22_-_38577782 | 0.26 |
ENST00000430886.1 ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr2_-_112614424 | 0.26 |
ENST00000427997.1 |
ANAPC1 |
anaphase promoting complex subunit 1 |
| chr11_-_75017734 | 0.26 |
ENST00000532525.1 |
ARRB1 |
arrestin, beta 1 |
| chr12_-_15114191 | 0.26 |
ENST00000541380.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr5_-_156390230 | 0.26 |
ENST00000407087.3 ENST00000274532.2 |
TIMD4 |
T-cell immunoglobulin and mucin domain containing 4 |
| chr15_+_68570062 | 0.26 |
ENST00000306917.4 |
FEM1B |
fem-1 homolog b (C. elegans) |
| chr3_+_184081175 | 0.25 |
ENST00000452961.1 ENST00000296223.3 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr7_-_75115548 | 0.25 |
ENST00000453279.2 |
POM121C |
POM121 transmembrane nucleoporin C |
| chr2_+_169312350 | 0.25 |
ENST00000305747.6 |
CERS6 |
ceramide synthase 6 |
| chr6_-_109330702 | 0.24 |
ENST00000356644.7 |
SESN1 |
sestrin 1 |
| chr18_+_61144160 | 0.24 |
ENST00000489441.1 ENST00000424602.1 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr2_+_71127699 | 0.24 |
ENST00000234392.2 |
VAX2 |
ventral anterior homeobox 2 |
| chr2_+_169312725 | 0.24 |
ENST00000392687.4 |
CERS6 |
ceramide synthase 6 |
| chr12_+_49687425 | 0.24 |
ENST00000257860.4 |
PRPH |
peripherin |
| chr14_+_39703112 | 0.24 |
ENST00000555143.1 ENST00000280082.3 |
MIA2 |
melanoma inhibitory activity 2 |
| chr22_-_38577731 | 0.24 |
ENST00000335539.3 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr1_+_46152886 | 0.23 |
ENST00000372025.4 |
TMEM69 |
transmembrane protein 69 |
| chr13_+_48611665 | 0.23 |
ENST00000258662.2 |
NUDT15 |
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr17_+_26662730 | 0.23 |
ENST00000226225.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr1_-_93426998 | 0.21 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr8_+_145218673 | 0.21 |
ENST00000326134.5 |
MROH1 |
maestro heat-like repeat family member 1 |
| chr6_+_55039050 | 0.21 |
ENST00000370862.3 |
HCRTR2 |
hypocretin (orexin) receptor 2 |
| chr11_+_62009653 | 0.21 |
ENST00000244926.3 |
SCGB1D2 |
secretoglobin, family 1D, member 2 |
| chr8_-_49833978 | 0.20 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr12_-_125348448 | 0.20 |
ENST00000339570.5 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr22_+_36044411 | 0.20 |
ENST00000409652.4 |
APOL6 |
apolipoprotein L, 6 |
| chr1_-_201398985 | 0.20 |
ENST00000336092.4 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr3_+_45017722 | 0.20 |
ENST00000265564.7 |
EXOSC7 |
exosome component 7 |
| chr3_+_184081137 | 0.20 |
ENST00000443489.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chrX_-_111923145 | 0.20 |
ENST00000371968.3 ENST00000536453.1 |
LHFPL1 |
lipoma HMGIC fusion partner-like 1 |
| chr3_+_37441062 | 0.20 |
ENST00000426078.1 |
C3orf35 |
chromosome 3 open reading frame 35 |
| chr12_-_125348329 | 0.20 |
ENST00000546215.1 ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr20_-_52790512 | 0.19 |
ENST00000216862.3 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr3_+_70048881 | 0.19 |
ENST00000483525.1 |
RP11-460N16.1 |
RP11-460N16.1 |
| chr2_+_47168630 | 0.19 |
ENST00000263737.6 |
TTC7A |
tetratricopeptide repeat domain 7A |
| chr8_+_22437965 | 0.19 |
ENST00000409141.1 ENST00000265810.4 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr8_-_117778494 | 0.19 |
ENST00000276682.4 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
| chr4_-_21546272 | 0.18 |
ENST00000509207.1 |
KCNIP4 |
Kv channel interacting protein 4 |
| chr4_+_8321882 | 0.18 |
ENST00000509453.1 ENST00000503186.1 |
RP11-774O3.2 RP11-774O3.1 |
RP11-774O3.2 RP11-774O3.1 |
| chr17_+_48556158 | 0.18 |
ENST00000258955.2 |
RSAD1 |
radical S-adenosyl methionine domain containing 1 |
| chr17_-_4090402 | 0.18 |
ENST00000574736.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chr2_+_234600253 | 0.18 |
ENST00000373424.1 ENST00000441351.1 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr16_+_10479906 | 0.18 |
ENST00000562527.1 ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2 |
activating transcription factor 7 interacting protein 2 |
| chr15_+_69857515 | 0.18 |
ENST00000559477.1 |
RP11-279F6.1 |
RP11-279F6.1 |
| chr19_+_4007644 | 0.18 |
ENST00000262971.2 |
PIAS4 |
protein inhibitor of activated STAT, 4 |
| chr11_+_46366918 | 0.17 |
ENST00000528615.1 ENST00000395574.3 |
DGKZ |
diacylglycerol kinase, zeta |
| chr10_-_101190202 | 0.17 |
ENST00000543866.1 ENST00000370508.5 |
GOT1 |
glutamic-oxaloacetic transaminase 1, soluble |
| chr19_-_52150053 | 0.17 |
ENST00000599649.1 ENST00000429354.3 ENST00000360844.6 ENST00000222107.4 |
SIGLEC5 SIGLEC14 SIGLEC5 |
SIGLEC5 sialic acid binding Ig-like lectin 14 sialic acid binding Ig-like lectin 5 |
| chr20_+_5731083 | 0.17 |
ENST00000445603.1 ENST00000442185.1 |
C20orf196 |
chromosome 20 open reading frame 196 |
| chr2_+_113299990 | 0.17 |
ENST00000537335.1 ENST00000417433.2 |
POLR1B |
polymerase (RNA) I polypeptide B, 128kDa |
| chr20_+_48807351 | 0.17 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr17_-_14140166 | 0.17 |
ENST00000420162.2 ENST00000431716.2 |
CDRT15 |
CMT1A duplicated region transcript 15 |
| chr19_-_47288162 | 0.17 |
ENST00000594991.1 |
SLC1A5 |
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr19_+_535835 | 0.17 |
ENST00000607527.1 ENST00000606065.1 |
CDC34 |
cell division cycle 34 |
| chr11_+_64073699 | 0.16 |
ENST00000405666.1 ENST00000468670.1 |
ESRRA |
estrogen-related receptor alpha |
| chr3_+_37440653 | 0.16 |
ENST00000328376.5 ENST00000452017.2 |
C3orf35 |
chromosome 3 open reading frame 35 |
| chr1_+_236558694 | 0.16 |
ENST00000359362.5 |
EDARADD |
EDAR-associated death domain |
| chr19_+_46498704 | 0.16 |
ENST00000595358.1 ENST00000594672.1 ENST00000536603.1 |
CCDC61 |
coiled-coil domain containing 61 |
| chr19_-_3500635 | 0.16 |
ENST00000250937.3 |
DOHH |
deoxyhypusine hydroxylase/monooxygenase |
| chr11_-_71823266 | 0.16 |
ENST00000538919.1 ENST00000539395.1 ENST00000542531.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr21_-_27462351 | 0.16 |
ENST00000448850.1 |
APP |
amyloid beta (A4) precursor protein |
| chr4_+_186317133 | 0.16 |
ENST00000507753.1 |
ANKRD37 |
ankyrin repeat domain 37 |
| chr9_+_139553306 | 0.15 |
ENST00000371699.1 |
EGFL7 |
EGF-like-domain, multiple 7 |
| chr3_-_50340996 | 0.15 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chr8_+_29953163 | 0.15 |
ENST00000518192.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr17_-_12921270 | 0.15 |
ENST00000578071.1 ENST00000426905.3 ENST00000395962.2 ENST00000583371.1 ENST00000338034.4 |
ELAC2 |
elaC ribonuclease Z 2 |
| chr4_-_7436671 | 0.15 |
ENST00000319098.4 |
PSAPL1 |
prosaposin-like 1 (gene/pseudogene) |
| chr2_+_47168313 | 0.14 |
ENST00000319190.5 ENST00000394850.2 ENST00000536057.1 |
TTC7A |
tetratricopeptide repeat domain 7A |
| chr14_+_58754751 | 0.14 |
ENST00000598233.1 |
AL132989.1 |
AL132989.1 |
| chr1_-_94312706 | 0.14 |
ENST00000370244.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr20_+_57264187 | 0.14 |
ENST00000525967.1 ENST00000525817.1 |
NPEPL1 |
aminopeptidase-like 1 |
| chr3_+_47844399 | 0.14 |
ENST00000446256.2 ENST00000445061.1 |
DHX30 |
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr11_+_36317830 | 0.14 |
ENST00000530639.1 |
PRR5L |
proline rich 5 like |
| chr4_+_3388057 | 0.14 |
ENST00000538395.1 |
RGS12 |
regulator of G-protein signaling 12 |
| chr5_+_132009675 | 0.14 |
ENST00000231449.2 ENST00000350025.2 |
IL4 |
interleukin 4 |
| chr7_-_99869799 | 0.14 |
ENST00000436886.2 |
GATS |
GATS, stromal antigen 3 opposite strand |
| chr20_+_816695 | 0.14 |
ENST00000246100.3 |
FAM110A |
family with sequence similarity 110, member A |
| chr5_+_180326077 | 0.14 |
ENST00000231229.4 ENST00000340184.4 ENST00000400707.3 |
BTNL8 |
butyrophilin-like 8 |
| chrX_+_72667090 | 0.13 |
ENST00000373514.2 |
CDX4 |
caudal type homeobox 4 |
| chr21_+_43823983 | 0.13 |
ENST00000291535.6 ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A |
ubiquitin associated and SH3 domain containing A |
| chr18_-_49557 | 0.13 |
ENST00000308911.6 |
RP11-683L23.1 |
Tubulin beta-8 chain-like protein LOC260334 |
| chr3_+_132316081 | 0.13 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr10_+_5726764 | 0.13 |
ENST00000328090.5 ENST00000496681.1 |
FAM208B |
family with sequence similarity 208, member B |
| chrX_-_129244655 | 0.13 |
ENST00000335997.7 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
| chr11_+_65122216 | 0.13 |
ENST00000309880.5 |
TIGD3 |
tigger transposable element derived 3 |
| chr22_+_29168652 | 0.12 |
ENST00000249064.4 ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117 |
coiled-coil domain containing 117 |
| chrX_-_45710920 | 0.12 |
ENST00000456532.1 |
RP5-1158E12.3 |
RP5-1158E12.3 |
| chr8_+_67341239 | 0.12 |
ENST00000320270.2 |
RRS1 |
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr1_-_228603694 | 0.12 |
ENST00000366697.2 |
TRIM17 |
tripartite motif containing 17 |
| chr17_-_42466864 | 0.12 |
ENST00000353281.4 ENST00000262407.5 |
ITGA2B |
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr1_+_155658849 | 0.12 |
ENST00000368336.5 ENST00000343043.3 ENST00000421487.2 ENST00000535183.1 ENST00000465375.1 ENST00000470830.1 |
DAP3 |
death associated protein 3 |
| chr1_-_150738261 | 0.12 |
ENST00000448301.2 ENST00000368985.3 |
CTSS |
cathepsin S |
| chr8_+_22853345 | 0.11 |
ENST00000522948.1 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
| chr17_-_37323318 | 0.11 |
ENST00000444555.1 |
ARL5C |
ADP-ribosylation factor-like 5C |
| chr1_-_20987982 | 0.11 |
ENST00000375048.3 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr1_-_46089718 | 0.11 |
ENST00000421127.2 ENST00000343901.2 ENST00000528266.1 |
CCDC17 |
coiled-coil domain containing 17 |
| chr5_+_72861560 | 0.11 |
ENST00000296792.4 ENST00000509005.1 ENST00000543251.1 ENST00000508686.1 ENST00000508491.1 |
UTP15 |
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
| chr13_+_27825446 | 0.11 |
ENST00000311549.6 |
RPL21 |
ribosomal protein L21 |
| chr1_-_38412683 | 0.11 |
ENST00000373024.3 ENST00000373023.2 |
INPP5B |
inositol polyphosphate-5-phosphatase, 75kDa |
| chr16_-_55909211 | 0.11 |
ENST00000520435.1 |
CES5A |
carboxylesterase 5A |
| chr2_+_68384976 | 0.11 |
ENST00000263657.2 |
PNO1 |
partner of NOB1 homolog (S. cerevisiae) |
| chr13_+_27825706 | 0.10 |
ENST00000272274.4 ENST00000319826.4 ENST00000326092.4 |
RPL21 |
ribosomal protein L21 |
| chr6_-_150212029 | 0.10 |
ENST00000529948.1 ENST00000357183.4 ENST00000367363.3 |
RAET1E |
retinoic acid early transcript 1E |
| chr5_+_170814803 | 0.10 |
ENST00000521672.1 ENST00000351986.6 ENST00000393820.2 ENST00000523622.1 |
NPM1 |
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| chr8_+_29952914 | 0.10 |
ENST00000321250.8 ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr16_-_55909255 | 0.10 |
ENST00000290567.9 |
CES5A |
carboxylesterase 5A |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.4 | 1.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 3.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.4 | 1.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.3 | 1.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.3 | 1.1 | GO:0042223 | response to molecule of fungal origin(GO:0002238) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.3 | 1.8 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 1.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.7 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.2 | 0.8 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.5 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 0.8 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.2 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.7 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.1 | 0.6 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 5.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 2.9 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 2.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 4.2 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 0.3 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.5 | GO:0090238 | regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 1.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.5 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.4 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.1 | 0.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.2 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.2 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.2 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0071288 | T-helper 1 cell lineage commitment(GO:0002296) B cell costimulation(GO:0031296) cellular response to mercury ion(GO:0071288) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.3 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 1.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.6 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 1.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.0 | 2.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.0 | GO:0060435 | branchiomeric skeletal muscle development(GO:0014707) bronchiole development(GO:0060435) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.0 | GO:0045659 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.2 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.1 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 2.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 1.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 1.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 4.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.8 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 2.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 5.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 2.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.4 | 1.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 1.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.6 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 2.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.6 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 2.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 4.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 1.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.2 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 1.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.2 | GO:0070546 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.4 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 2.9 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.5 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 2.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 1.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 4.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |