Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for UGGCACU
Z-value: 0.73
Activity profile of UGGCACU motif
Sorted Z-values of UGGCACU motif
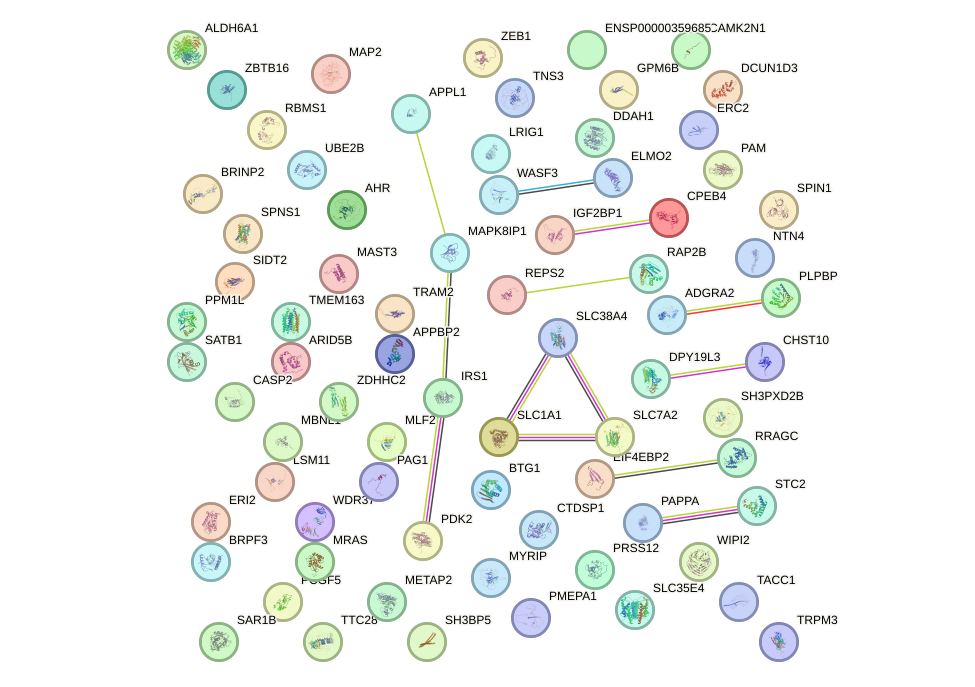
Network of associatons between targets according to the STRING database.
First level regulatory network of UGGCACU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_31608054 | 0.70 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr11_+_113930291 | 0.68 |
ENST00000335953.4 |
ZBTB16 |
zinc finger and BTB domain containing 16 |
| chrX_+_16964794 | 0.67 |
ENST00000357277.3 |
REPS2 |
RALBP1 associated Eps domain containing 2 |
| chr9_+_4490394 | 0.41 |
ENST00000262352.3 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr16_-_80838195 | 0.38 |
ENST00000570137.2 |
CDYL2 |
chromodomain protein, Y-like 2 |
| chr7_-_47621736 | 0.37 |
ENST00000311160.9 |
TNS3 |
tensin 3 |
| chr3_+_69812877 | 0.34 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr8_+_37654424 | 0.34 |
ENST00000315215.7 |
GPR124 |
G protein-coupled receptor 124 |
| chr9_+_79074068 | 0.32 |
ENST00000444201.2 ENST00000376730.4 |
GCNT1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr5_-_172756506 | 0.32 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr11_-_82782861 | 0.32 |
ENST00000524635.1 ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30 |
RAB30, member RAS oncogene family |
| chr12_-_92539614 | 0.31 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr12_-_96184533 | 0.30 |
ENST00000343702.4 ENST00000344911.4 |
NTN4 |
netrin 4 |
| chrX_+_9431324 | 0.26 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr6_-_57087042 | 0.26 |
ENST00000317483.3 |
RAB23 |
RAB23, member RAS oncogene family |
| chr17_+_47074758 | 0.26 |
ENST00000290341.3 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
| chr5_+_67511524 | 0.26 |
ENST00000521381.1 ENST00000521657.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr8_+_38614807 | 0.25 |
ENST00000330691.6 ENST00000348567.4 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr8_+_17013515 | 0.23 |
ENST00000262096.8 |
ZDHHC2 |
zinc finger, DHHC-type containing 2 |
| chr10_+_92980517 | 0.23 |
ENST00000336126.5 |
PCGF5 |
polycomb group ring finger 5 |
| chr3_+_138066539 | 0.23 |
ENST00000289104.4 |
MRAS |
muscle RAS oncogene homolog |
| chr8_+_77593448 | 0.21 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr9_+_91003271 | 0.21 |
ENST00000375859.3 ENST00000541629.1 |
SPIN1 |
spindlin 1 |
| chr16_+_8768422 | 0.20 |
ENST00000268251.8 ENST00000564714.1 |
ABAT |
4-aminobutyrate aminotransferase |
| chr2_+_210444142 | 0.20 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr2_-_227664474 | 0.19 |
ENST00000305123.5 |
IRS1 |
insulin receptor substrate 1 |
| chr3_-_9994021 | 0.18 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chr1_-_86043921 | 0.18 |
ENST00000535924.2 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr7_+_142985308 | 0.18 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr3_-_66551351 | 0.17 |
ENST00000273261.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr5_+_133706865 | 0.17 |
ENST00000265339.2 |
UBE2B |
ubiquitin-conjugating enzyme E2B |
| chr8_+_37620097 | 0.15 |
ENST00000328195.3 ENST00000523358.1 ENST00000523187.1 |
PROSC |
proline synthetase co-transcribed homolog (bacterial) |
| chr12_+_60083118 | 0.15 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chrX_-_106959631 | 0.15 |
ENST00000486554.1 ENST00000372390.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr4_+_77870856 | 0.15 |
ENST00000264893.6 ENST00000502584.1 ENST00000510641.1 |
SEPT11 |
septin 11 |
| chr5_+_157170703 | 0.14 |
ENST00000286307.5 |
LSM11 |
LSM11, U7 small nuclear RNA associated |
| chr12_-_47219733 | 0.14 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chr20_-_56284816 | 0.13 |
ENST00000395819.3 ENST00000341744.3 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr19_+_32896697 | 0.13 |
ENST00000586987.1 |
DPY19L3 |
dpy-19-like 3 (C. elegans) |
| chr6_-_34360413 | 0.13 |
ENST00000607016.1 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
| chr5_+_173315283 | 0.12 |
ENST00000265085.5 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
| chr22_+_30792846 | 0.12 |
ENST00000312932.9 ENST00000428195.1 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr13_+_27131887 | 0.12 |
ENST00000335327.5 |
WASF3 |
WAS protein family, member 3 |
| chr17_+_48172639 | 0.12 |
ENST00000503176.1 ENST00000503614.1 |
PDK2 |
pyruvate dehydrogenase kinase, isozyme 2 |
| chr2_+_219264466 | 0.12 |
ENST00000273062.2 |
CTDSP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr10_+_72164135 | 0.12 |
ENST00000373218.4 |
EIF4EBP2 |
eukaryotic translation initiation factor 4E binding protein 2 |
| chr10_+_1095416 | 0.12 |
ENST00000358220.1 |
WDR37 |
WD repeat domain 37 |
| chr6_-_52441713 | 0.11 |
ENST00000182527.3 |
TRAM2 |
translocation associated membrane protein 2 |
| chr10_+_63661053 | 0.11 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr8_+_17354587 | 0.11 |
ENST00000494857.1 ENST00000522656.1 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr3_+_39851094 | 0.11 |
ENST00000302541.6 |
MYRIP |
myosin VIIA and Rab interacting protein |
| chr1_+_112162381 | 0.11 |
ENST00000433097.1 ENST00000369709.3 ENST00000436150.2 |
RAP1A |
RAP1A, member of RAS oncogene family |
| chr1_-_39325431 | 0.11 |
ENST00000373001.3 |
RRAGC |
Ras-related GTP binding C |
| chr22_+_31031639 | 0.11 |
ENST00000343605.4 ENST00000300385.8 |
SLC35E4 |
solute carrier family 35, member E4 |
| chr17_+_66508537 | 0.10 |
ENST00000392711.1 ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr9_+_118916082 | 0.10 |
ENST00000328252.3 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr20_+_11871371 | 0.10 |
ENST00000254977.3 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr4_-_119274121 | 0.10 |
ENST00000296498.3 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
| chr3_+_191046810 | 0.10 |
ENST00000392455.3 ENST00000392456.3 |
CCDC50 |
coiled-coil domain containing 50 |
| chr5_-_133968529 | 0.10 |
ENST00000402673.2 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
| chr3_-_15374033 | 0.10 |
ENST00000253688.5 ENST00000383791.3 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
| chrX_-_13956737 | 0.10 |
ENST00000454189.2 |
GPM6B |
glycoprotein M6B |
| chr14_-_53619816 | 0.10 |
ENST00000323669.5 ENST00000395606.1 ENST00000357758.3 |
DDHD1 |
DDHD domain containing 1 |
| chr3_+_152017181 | 0.09 |
ENST00000498502.1 ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chrX_+_12993202 | 0.09 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr1_-_20812690 | 0.09 |
ENST00000375078.3 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr3_+_57261743 | 0.09 |
ENST00000288266.3 |
APPL1 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr20_-_45035223 | 0.09 |
ENST00000450812.1 ENST00000290246.6 ENST00000439931.2 ENST00000396391.1 |
ELMO2 |
engulfment and cell motility 2 |
| chr3_+_160473996 | 0.09 |
ENST00000498165.1 |
PPM1L |
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr2_-_135476552 | 0.09 |
ENST00000281924.6 |
TMEM163 |
transmembrane protein 163 |
| chr17_-_58603568 | 0.09 |
ENST00000083182.3 |
APPBP2 |
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr14_-_74551172 | 0.08 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr3_-_56502375 | 0.08 |
ENST00000288221.6 |
ERC2 |
ELKS/RAB6-interacting/CAST family member 2 |
| chr15_-_83316254 | 0.08 |
ENST00000567678.1 ENST00000450751.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr7_+_17338239 | 0.08 |
ENST00000242057.4 |
AHR |
aryl hydrocarbon receptor |
| chr17_-_42908155 | 0.08 |
ENST00000426548.1 ENST00000590758.1 ENST00000591424.1 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
| chr11_+_45907177 | 0.08 |
ENST00000241014.2 |
MAPK8IP1 |
mitogen-activated protein kinase 8 interacting protein 1 |
| chr2_-_101034070 | 0.08 |
ENST00000264249.3 |
CHST10 |
carbohydrate sulfotransferase 10 |
| chr5_+_102201430 | 0.08 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr5_-_171881491 | 0.08 |
ENST00000311601.5 |
SH3PXD2B |
SH3 and PX domains 2B |
| chr19_+_18208603 | 0.08 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr7_+_7811992 | 0.08 |
ENST00000406829.1 |
RPA3-AS1 |
RPA3 antisense RNA 1 |
| chr15_-_75744014 | 0.08 |
ENST00000394947.3 ENST00000565264.1 |
SIN3A |
SIN3 transcription regulator family member A |
| chr3_+_152879985 | 0.08 |
ENST00000323534.2 |
RAP2B |
RAP2B, member of RAS oncogene family |
| chr1_+_177140633 | 0.08 |
ENST00000361539.4 |
BRINP2 |
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr8_-_82024290 | 0.08 |
ENST00000220597.4 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr3_-_18466787 | 0.08 |
ENST00000338745.6 ENST00000450898.1 |
SATB1 |
SATB homeobox 1 |
| chr22_-_29075853 | 0.08 |
ENST00000397906.2 |
TTC28 |
tetratricopeptide repeat domain 28 |
| chr9_-_73736511 | 0.08 |
ENST00000377110.3 ENST00000377111.2 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr11_+_117049445 | 0.08 |
ENST00000324225.4 ENST00000532960.1 |
SIDT2 |
SID1 transmembrane family, member 2 |
| chr2_-_161350305 | 0.08 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr7_+_5229819 | 0.07 |
ENST00000288828.4 ENST00000401525.3 ENST00000404704.3 |
WIPI2 |
WD repeat domain, phosphoinositide interacting 2 |
| chr6_+_64281906 | 0.07 |
ENST00000370651.3 |
PTP4A1 |
protein tyrosine phosphatase type IVA, member 1 |
| chr6_+_36164487 | 0.07 |
ENST00000357641.6 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
| chr16_-_20911641 | 0.07 |
ENST00000564349.1 ENST00000324344.4 |
ERI2 DCUN1D3 |
ERI1 exoribonuclease family member 2 DCN1, defective in cullin neddylation 1, domain containing 3 |
| chr2_+_45878790 | 0.07 |
ENST00000306156.3 |
PRKCE |
protein kinase C, epsilon |
| chr1_+_113933581 | 0.07 |
ENST00000307546.9 ENST00000369615.1 ENST00000369611.4 |
MAGI3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chrX_-_83442915 | 0.07 |
ENST00000262752.2 ENST00000543399.1 |
RPS6KA6 |
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr12_+_27396901 | 0.07 |
ENST00000541191.1 ENST00000389032.3 |
STK38L |
serine/threonine kinase 38 like |
| chr5_+_76506706 | 0.07 |
ENST00000340978.3 ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B |
phosphodiesterase 8B |
| chr13_-_23949671 | 0.07 |
ENST00000402364.1 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr7_-_28220354 | 0.07 |
ENST00000283928.5 |
JAZF1 |
JAZF zinc finger 1 |
| chr11_-_64612041 | 0.07 |
ENST00000342711.5 |
CDC42BPG |
CDC42 binding protein kinase gamma (DMPK-like) |
| chr9_+_2015335 | 0.07 |
ENST00000349721.2 ENST00000357248.2 ENST00000450198.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_-_114246635 | 0.07 |
ENST00000338205.5 |
KIAA0368 |
KIAA0368 |
| chr18_+_63418068 | 0.06 |
ENST00000397968.2 |
CDH7 |
cadherin 7, type 2 |
| chr1_+_118148556 | 0.06 |
ENST00000369448.3 |
FAM46C |
family with sequence similarity 46, member C |
| chr13_+_49550015 | 0.06 |
ENST00000492622.2 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr8_-_57123815 | 0.06 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr19_+_36630961 | 0.06 |
ENST00000587718.1 ENST00000592483.1 ENST00000590874.1 ENST00000588815.1 |
CAPNS1 |
calpain, small subunit 1 |
| chr18_+_67956135 | 0.06 |
ENST00000397942.3 |
SOCS6 |
suppressor of cytokine signaling 6 |
| chr17_+_61699766 | 0.06 |
ENST00000579585.1 ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3 |
mitogen-activated protein kinase kinase kinase 3 |
| chr7_+_98246588 | 0.06 |
ENST00000265634.3 |
NPTX2 |
neuronal pentraxin II |
| chr11_-_45687128 | 0.06 |
ENST00000308064.2 |
CHST1 |
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr6_+_52226897 | 0.06 |
ENST00000442253.2 |
PAQR8 |
progestin and adipoQ receptor family member VIII |
| chr14_+_55034599 | 0.06 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chrX_+_110339439 | 0.06 |
ENST00000372010.1 ENST00000519681.1 ENST00000372007.5 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr11_+_120207787 | 0.06 |
ENST00000397843.2 ENST00000356641.3 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr9_+_100263912 | 0.06 |
ENST00000259365.4 |
TMOD1 |
tropomodulin 1 |
| chr12_-_118541743 | 0.06 |
ENST00000359236.5 |
VSIG10 |
V-set and immunoglobulin domain containing 10 |
| chr5_-_168728103 | 0.06 |
ENST00000519560.1 |
SLIT3 |
slit homolog 3 (Drosophila) |
| chr1_-_68299130 | 0.06 |
ENST00000370982.3 |
GNG12 |
guanine nucleotide binding protein (G protein), gamma 12 |
| chrX_+_72783026 | 0.06 |
ENST00000373504.6 ENST00000373502.5 |
CHIC1 |
cysteine-rich hydrophobic domain 1 |
| chr2_+_205410516 | 0.06 |
ENST00000406610.2 ENST00000462231.1 |
PARD3B |
par-3 family cell polarity regulator beta |
| chr4_-_114682936 | 0.05 |
ENST00000454265.2 ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr13_-_31038370 | 0.05 |
ENST00000399489.1 ENST00000339872.4 |
HMGB1 |
high mobility group box 1 |
| chr5_-_171433819 | 0.05 |
ENST00000296933.6 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr16_+_56672571 | 0.05 |
ENST00000290705.8 |
MT1A |
metallothionein 1A |
| chr14_+_55518349 | 0.05 |
ENST00000395468.4 |
MAPK1IP1L |
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr3_-_39195037 | 0.05 |
ENST00000273153.5 |
CSRNP1 |
cysteine-serine-rich nuclear protein 1 |
| chr12_+_69864129 | 0.05 |
ENST00000547219.1 ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
| chr9_+_32384617 | 0.05 |
ENST00000379923.1 ENST00000309951.6 ENST00000541043.1 |
ACO1 |
aconitase 1, soluble |
| chr3_-_15901278 | 0.05 |
ENST00000399451.2 |
ANKRD28 |
ankyrin repeat domain 28 |
| chr19_+_3094398 | 0.05 |
ENST00000078429.4 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
| chr11_-_6677018 | 0.05 |
ENST00000299441.3 |
DCHS1 |
dachsous cadherin-related 1 |
| chr7_-_27183263 | 0.05 |
ENST00000222726.3 |
HOXA5 |
homeobox A5 |
| chr20_+_277737 | 0.05 |
ENST00000382352.3 |
ZCCHC3 |
zinc finger, CCHC domain containing 3 |
| chr1_+_24286287 | 0.05 |
ENST00000334351.7 ENST00000374468.1 |
PNRC2 |
proline-rich nuclear receptor coactivator 2 |
| chr1_+_7831323 | 0.05 |
ENST00000054666.6 |
VAMP3 |
vesicle-associated membrane protein 3 |
| chr2_+_20646824 | 0.05 |
ENST00000272233.4 |
RHOB |
ras homolog family member B |
| chr2_+_204192942 | 0.05 |
ENST00000295851.5 ENST00000261017.5 |
ABI2 |
abl-interactor 2 |
| chr9_-_139922631 | 0.05 |
ENST00000341511.6 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr5_+_149887672 | 0.05 |
ENST00000261797.6 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr12_-_26986076 | 0.05 |
ENST00000381340.3 |
ITPR2 |
inositol 1,4,5-trisphosphate receptor, type 2 |
| chr2_+_173600671 | 0.04 |
ENST00000409036.1 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr15_-_51058005 | 0.04 |
ENST00000261854.5 |
SPPL2A |
signal peptide peptidase like 2A |
| chr2_+_121010324 | 0.04 |
ENST00000272519.5 |
RALB |
v-ral simian leukemia viral oncogene homolog B |
| chr7_+_106809406 | 0.04 |
ENST00000468410.1 ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1 |
HMG-box transcription factor 1 |
| chr4_-_78740511 | 0.04 |
ENST00000504123.1 ENST00000264903.4 ENST00000515441.1 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
| chr1_-_154943212 | 0.04 |
ENST00000368445.5 ENST00000448116.2 ENST00000368449.4 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr16_+_77822427 | 0.04 |
ENST00000302536.2 |
VAT1L |
vesicle amine transport 1-like |
| chr6_-_159239257 | 0.04 |
ENST00000337147.7 ENST00000392177.4 |
EZR |
ezrin |
| chr3_-_72496035 | 0.04 |
ENST00000477973.2 |
RYBP |
RING1 and YY1 binding protein |
| chr1_-_47134101 | 0.04 |
ENST00000576409.1 |
ATPAF1 |
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr3_+_89156674 | 0.04 |
ENST00000336596.2 |
EPHA3 |
EPH receptor A3 |
| chr17_+_30677136 | 0.04 |
ENST00000394670.4 ENST00000321233.6 ENST00000394673.2 ENST00000341711.6 ENST00000579634.1 ENST00000580759.1 ENST00000342555.6 ENST00000577908.1 ENST00000394679.5 ENST00000582165.1 |
ZNF207 |
zinc finger protein 207 |
| chr12_-_54673871 | 0.04 |
ENST00000209875.4 |
CBX5 |
chromobox homolog 5 |
| chr15_+_54305101 | 0.04 |
ENST00000260323.11 ENST00000545554.1 ENST00000537900.1 |
UNC13C |
unc-13 homolog C (C. elegans) |
| chr12_-_58027138 | 0.04 |
ENST00000341156.4 |
B4GALNT1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr5_+_131705438 | 0.04 |
ENST00000245407.3 |
SLC22A5 |
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr2_-_44588893 | 0.04 |
ENST00000409272.1 ENST00000410081.1 ENST00000541738.1 |
PREPL |
prolyl endopeptidase-like |
| chr12_-_76953284 | 0.04 |
ENST00000547544.1 ENST00000393249.2 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr20_-_32262165 | 0.03 |
ENST00000606690.1 ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3 |
N-terminal EF-hand calcium binding protein 3 |
| chr12_+_110906169 | 0.03 |
ENST00000377673.5 |
FAM216A |
family with sequence similarity 216, member A |
| chr10_+_102821551 | 0.03 |
ENST00000370200.5 |
KAZALD1 |
Kazal-type serine peptidase inhibitor domain 1 |
| chr10_-_15413035 | 0.03 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chr9_-_4741255 | 0.03 |
ENST00000381809.3 |
AK3 |
adenylate kinase 3 |
| chr8_+_42752053 | 0.03 |
ENST00000307602.4 |
HOOK3 |
hook microtubule-tethering protein 3 |
| chr4_+_48343339 | 0.03 |
ENST00000264313.6 |
SLAIN2 |
SLAIN motif family, member 2 |
| chr7_+_140774032 | 0.03 |
ENST00000565468.1 |
TMEM178B |
transmembrane protein 178B |
| chr1_+_9352939 | 0.03 |
ENST00000328089.6 |
SPSB1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
| chrX_+_154997474 | 0.03 |
ENST00000302805.2 |
SPRY3 |
sprouty homolog 3 (Drosophila) |
| chr5_+_65222299 | 0.03 |
ENST00000284037.5 |
ERBB2IP |
erbb2 interacting protein |
| chr7_+_107301065 | 0.03 |
ENST00000265715.3 |
SLC26A4 |
solute carrier family 26 (anion exchanger), member 4 |
| chr20_-_3996036 | 0.03 |
ENST00000336095.6 |
RNF24 |
ring finger protein 24 |
| chr20_+_32581452 | 0.03 |
ENST00000375114.3 ENST00000448364.1 |
RALY |
RALY heterogeneous nuclear ribonucleoprotein |
| chr11_-_88796803 | 0.03 |
ENST00000418177.2 ENST00000455756.2 |
GRM5 |
glutamate receptor, metabotropic 5 |
| chrX_-_19905703 | 0.03 |
ENST00000397821.3 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr5_+_17217669 | 0.03 |
ENST00000322611.3 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
| chr3_-_141868357 | 0.03 |
ENST00000489671.1 ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_147124547 | 0.03 |
ENST00000491672.1 ENST00000383075.3 |
ZIC4 |
Zic family member 4 |
| chr7_+_100728720 | 0.03 |
ENST00000306085.6 ENST00000412507.1 |
TRIM56 |
tripartite motif containing 56 |
| chr11_-_77532050 | 0.03 |
ENST00000308488.6 |
RSF1 |
remodeling and spacing factor 1 |
| chr12_+_93771659 | 0.03 |
ENST00000337179.5 ENST00000415493.2 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr2_+_46844290 | 0.03 |
ENST00000238892.3 |
CRIPT |
cysteine-rich PDZ-binding protein |
| chr1_-_55352834 | 0.03 |
ENST00000371269.3 |
DHCR24 |
24-dehydrocholesterol reductase |
| chrX_-_102942961 | 0.03 |
ENST00000434230.1 ENST00000418819.1 ENST00000360458.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr19_+_11466062 | 0.03 |
ENST00000251473.5 ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410 |
Lipid phosphate phosphatase-related protein type 2 |
| chr1_-_153895377 | 0.03 |
ENST00000368655.4 |
GATAD2B |
GATA zinc finger domain containing 2B |
| chr9_-_35732362 | 0.03 |
ENST00000314888.9 ENST00000540444.1 |
TLN1 |
talin 1 |
| chr1_+_198126093 | 0.03 |
ENST00000367385.4 ENST00000442588.1 ENST00000538004.1 |
NEK7 |
NIMA-related kinase 7 |
| chr1_+_192778161 | 0.03 |
ENST00000235382.5 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
| chr15_+_79165151 | 0.03 |
ENST00000331268.5 |
MORF4L1 |
mortality factor 4 like 1 |
| chr8_-_124553437 | 0.03 |
ENST00000517956.1 ENST00000443022.2 |
FBXO32 |
F-box protein 32 |
| chr5_+_143584814 | 0.03 |
ENST00000507359.3 |
KCTD16 |
potassium channel tetramerization domain containing 16 |
| chr1_+_36273743 | 0.03 |
ENST00000373210.3 |
AGO4 |
argonaute RISC catalytic component 4 |
| chr5_+_139493665 | 0.03 |
ENST00000331327.3 |
PURA |
purine-rich element binding protein A |
| chr6_+_149887377 | 0.03 |
ENST00000367419.5 |
GINM1 |
glycoprotein integral membrane 1 |
| chr18_-_23670546 | 0.03 |
ENST00000542743.1 ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18 |
synovial sarcoma translocation, chromosome 18 |
| chr3_-_44519131 | 0.03 |
ENST00000425708.2 ENST00000396077.2 |
ZNF445 |
zinc finger protein 445 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.4 | GO:0070777 | sulfur amino acid transport(GO:0000101) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0034226 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.2 | GO:1901910 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0005287 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0052841 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0051538 | iron-responsive element binding(GO:0030350) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.0 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |