Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for VAX1_GSX2
Z-value: 0.14
Transcription factors associated with VAX1_GSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VAX1
|
ENSG00000148704.8 | VAX1 |
|
GSX2
|
ENSG00000180613.6 | GSX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GSX2 | hg19_v2_chr4_+_54966198_54966340 | -0.52 | 1.8e-01 | Click! |
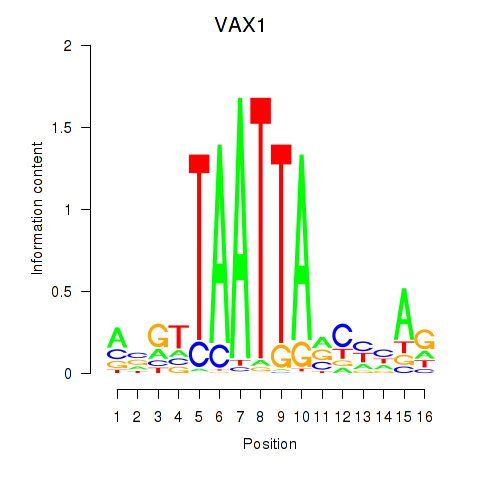
Activity profile of VAX1_GSX2 motif
Sorted Z-values of VAX1_GSX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX1_GSX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_138453606 | 0.31 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr5_-_20575959 | 0.28 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr7_-_92777606 | 0.21 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr4_-_138453559 | 0.21 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr16_+_53133070 | 0.16 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr1_+_192778161 | 0.15 |
ENST00000235382.5 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
| chr17_-_66951474 | 0.14 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr2_-_188419078 | 0.13 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr20_-_50418947 | 0.12 |
ENST00000371539.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr20_-_50419055 | 0.12 |
ENST00000217086.4 |
SALL4 |
spalt-like transcription factor 4 |
| chr20_-_50418972 | 0.12 |
ENST00000395997.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr7_+_99717230 | 0.12 |
ENST00000262932.3 |
CNPY4 |
canopy FGF signaling regulator 4 |
| chr12_+_15699286 | 0.11 |
ENST00000442921.2 ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr22_-_32651326 | 0.11 |
ENST00000266086.4 |
SLC5A4 |
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr3_-_191000172 | 0.11 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr3_+_157154578 | 0.10 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr15_+_62853562 | 0.10 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr3_-_112565703 | 0.09 |
ENST00000488794.1 |
CD200R1L |
CD200 receptor 1-like |
| chr19_+_50016610 | 0.09 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr4_+_70894130 | 0.09 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr4_-_119759795 | 0.09 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr15_-_37393406 | 0.08 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr17_+_59489112 | 0.08 |
ENST00000335108.2 |
C17orf82 |
chromosome 17 open reading frame 82 |
| chr9_+_90112767 | 0.08 |
ENST00000408954.3 |
DAPK1 |
death-associated protein kinase 1 |
| chr12_+_26348246 | 0.08 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr9_-_95166841 | 0.08 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr3_-_105588231 | 0.07 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr9_+_90112590 | 0.07 |
ENST00000472284.1 |
DAPK1 |
death-associated protein kinase 1 |
| chr9_-_95166884 | 0.07 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr9_-_16728161 | 0.07 |
ENST00000603713.1 ENST00000603313.1 |
BNC2 |
basonuclin 2 |
| chr4_-_66536057 | 0.07 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr2_+_210517895 | 0.07 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr7_+_100770328 | 0.06 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr8_-_42623747 | 0.06 |
ENST00000534622.1 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr12_-_10607084 | 0.06 |
ENST00000408006.3 ENST00000544822.1 ENST00000536188.1 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr11_+_27015628 | 0.06 |
ENST00000318627.2 |
FIBIN |
fin bud initiation factor homolog (zebrafish) |
| chr11_-_26593649 | 0.06 |
ENST00000455601.2 |
MUC15 |
mucin 15, cell surface associated |
| chr3_+_108541545 | 0.06 |
ENST00000295756.6 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr11_-_26593677 | 0.05 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr1_+_36789335 | 0.05 |
ENST00000373137.2 |
RP11-268J15.5 |
RP11-268J15.5 |
| chr4_+_41614720 | 0.05 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr14_+_100485712 | 0.05 |
ENST00000544450.2 |
EVL |
Enah/Vasp-like |
| chr12_-_10605929 | 0.05 |
ENST00000347831.5 ENST00000359151.3 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr6_+_29274403 | 0.05 |
ENST00000377160.2 |
OR14J1 |
olfactory receptor, family 14, subfamily J, member 1 |
| chr18_-_54305658 | 0.05 |
ENST00000586262.1 ENST00000217515.6 |
TXNL1 |
thioredoxin-like 1 |
| chr1_+_186369704 | 0.05 |
ENST00000574641.1 |
OCLM |
oculomedin |
| chr11_-_26593779 | 0.05 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr19_+_45417921 | 0.05 |
ENST00000252491.4 ENST00000592885.1 ENST00000589781.1 |
APOC1 |
apolipoprotein C-I |
| chr7_-_14029283 | 0.05 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr5_-_150473127 | 0.04 |
ENST00000521001.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr3_+_108541608 | 0.04 |
ENST00000426646.1 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr7_-_81399411 | 0.04 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr9_+_12775011 | 0.04 |
ENST00000319264.3 |
LURAP1L |
leucine rich adaptor protein 1-like |
| chr1_+_154401791 | 0.04 |
ENST00000476006.1 |
IL6R |
interleukin 6 receptor |
| chr15_+_41913690 | 0.04 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chrX_+_43515467 | 0.04 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr3_-_164796269 | 0.04 |
ENST00000264382.3 |
SI |
sucrase-isomaltase (alpha-glucosidase) |
| chr5_-_147286065 | 0.04 |
ENST00000318315.4 ENST00000515291.1 |
C5orf46 |
chromosome 5 open reading frame 46 |
| chr19_-_58951496 | 0.04 |
ENST00000254166.3 |
ZNF132 |
zinc finger protein 132 |
| chr22_+_23154239 | 0.04 |
ENST00000390315.2 |
IGLV3-10 |
immunoglobulin lambda variable 3-10 |
| chr11_+_100862811 | 0.04 |
ENST00000303130.2 |
TMEM133 |
transmembrane protein 133 |
| chr3_-_105587879 | 0.04 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr3_+_111393501 | 0.04 |
ENST00000393934.3 |
PLCXD2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr14_+_22465771 | 0.03 |
ENST00000390445.2 |
TRAV17 |
T cell receptor alpha variable 17 |
| chr11_-_124981475 | 0.03 |
ENST00000532156.1 ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218 |
transmembrane protein 218 |
| chr13_-_36050819 | 0.03 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr12_-_10282681 | 0.03 |
ENST00000533022.1 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr2_+_105050794 | 0.03 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr13_+_36050881 | 0.03 |
ENST00000537702.1 |
NBEA |
neurobeachin |
| chr10_-_4285923 | 0.03 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr11_-_27722021 | 0.03 |
ENST00000356660.4 ENST00000418212.1 ENST00000533246.1 |
BDNF |
brain-derived neurotrophic factor |
| chr9_+_125133315 | 0.03 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_+_26348429 | 0.03 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr3_+_138340067 | 0.03 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr3_+_158519654 | 0.03 |
ENST00000415822.2 ENST00000392813.4 ENST00000264266.8 |
MFSD1 |
major facilitator superfamily domain containing 1 |
| chr11_-_65149422 | 0.03 |
ENST00000526432.1 ENST00000527174.1 |
SLC25A45 |
solute carrier family 25, member 45 |
| chr15_+_89631381 | 0.03 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr21_+_17792672 | 0.03 |
ENST00000602620.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr14_-_90085458 | 0.03 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr10_-_92681033 | 0.03 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr6_-_87804815 | 0.03 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr1_+_53925063 | 0.03 |
ENST00000371445.3 |
DMRTB1 |
DMRT-like family B with proline-rich C-terminal, 1 |
| chr11_-_96076334 | 0.03 |
ENST00000524717.1 |
MAML2 |
mastermind-like 2 (Drosophila) |
| chr7_-_121944491 | 0.03 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr17_-_18430160 | 0.03 |
ENST00000392176.3 |
FAM106A |
family with sequence similarity 106, member A |
| chr22_+_24990746 | 0.03 |
ENST00000456869.1 ENST00000411974.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr11_-_33913708 | 0.03 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr11_+_66276550 | 0.03 |
ENST00000419755.3 |
CTD-3074O7.11 |
Bardet-Biedl syndrome 1 protein |
| chr4_-_164534657 | 0.03 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr7_-_88425025 | 0.03 |
ENST00000297203.2 |
C7orf62 |
chromosome 7 open reading frame 62 |
| chr17_+_20978854 | 0.03 |
ENST00000456235.1 |
AC087393.1 |
AC087393.1 |
| chr1_+_117544366 | 0.03 |
ENST00000256652.4 ENST00000369470.1 |
CD101 |
CD101 molecule |
| chr12_+_18414446 | 0.03 |
ENST00000433979.1 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr15_+_64680003 | 0.03 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr12_+_113587558 | 0.03 |
ENST00000335621.6 |
CCDC42B |
coiled-coil domain containing 42B |
| chr18_-_67624160 | 0.03 |
ENST00000581982.1 ENST00000280200.4 |
CD226 |
CD226 molecule |
| chr1_-_68698222 | 0.03 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr10_-_135379132 | 0.03 |
ENST00000343131.5 |
SYCE1 |
synaptonemal complex central element protein 1 |
| chr3_-_99569821 | 0.03 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr11_-_13517565 | 0.03 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr2_-_73520667 | 0.03 |
ENST00000545030.1 ENST00000436467.2 |
EGR4 |
early growth response 4 |
| chrX_-_13835147 | 0.03 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr10_-_5046042 | 0.03 |
ENST00000421196.3 ENST00000455190.1 |
AKR1C2 |
aldo-keto reductase family 1, member C2 |
| chr15_+_89631647 | 0.02 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr7_-_99716952 | 0.02 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr10_+_5005598 | 0.02 |
ENST00000442997.1 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr17_-_72772462 | 0.02 |
ENST00000582870.1 ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9 |
N-acetyltransferase 9 (GCN5-related, putative) |
| chr14_+_37641012 | 0.02 |
ENST00000556667.1 |
SLC25A21-AS1 |
SLC25A21 antisense RNA 1 |
| chr1_+_144989309 | 0.02 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr3_+_62304712 | 0.02 |
ENST00000494481.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr1_+_214161854 | 0.02 |
ENST00000435016.1 |
PROX1 |
prospero homeobox 1 |
| chr3_+_138340049 | 0.02 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr15_+_84115868 | 0.02 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr10_-_28571015 | 0.02 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr11_+_33061543 | 0.02 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr10_+_47894023 | 0.02 |
ENST00000358474.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr7_-_14028488 | 0.02 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr19_+_42055879 | 0.02 |
ENST00000407170.2 ENST00000601116.1 ENST00000595395.1 |
CEACAM21 AC006129.2 |
carcinoembryonic antigen-related cell adhesion molecule 21 AC006129.2 |
| chr9_+_131062367 | 0.02 |
ENST00000601297.1 |
AL359091.2 |
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr1_-_36789755 | 0.02 |
ENST00000270824.1 |
EVA1B |
eva-1 homolog B (C. elegans) |
| chr1_-_150669604 | 0.02 |
ENST00000427665.1 ENST00000540514.1 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr6_+_29429217 | 0.02 |
ENST00000396792.2 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr13_-_41593425 | 0.02 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr1_+_81771806 | 0.02 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr6_+_26440700 | 0.02 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr1_-_205325850 | 0.02 |
ENST00000537168.1 |
KLHDC8A |
kelch domain containing 8A |
| chr7_-_99717463 | 0.02 |
ENST00000437822.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr12_+_10163231 | 0.02 |
ENST00000396502.1 ENST00000338896.5 |
CLEC12B |
C-type lectin domain family 12, member B |
| chr3_+_2933893 | 0.02 |
ENST00000397459.2 |
CNTN4 |
contactin 4 |
| chr4_+_78432907 | 0.02 |
ENST00000286758.4 |
CXCL13 |
chemokine (C-X-C motif) ligand 13 |
| chr1_-_152386732 | 0.02 |
ENST00000271835.3 |
CRNN |
cornulin |
| chr1_-_109618566 | 0.02 |
ENST00000338366.5 |
TAF13 |
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr16_-_31085514 | 0.02 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr2_-_3521518 | 0.02 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chr2_-_40680578 | 0.02 |
ENST00000455476.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr11_-_133826852 | 0.02 |
ENST00000533871.2 ENST00000321016.8 |
IGSF9B |
immunoglobulin superfamily, member 9B |
| chr7_+_120628731 | 0.02 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chrX_-_16887963 | 0.02 |
ENST00000380084.4 |
RBBP7 |
retinoblastoma binding protein 7 |
| chr11_+_5710919 | 0.02 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr3_+_28390637 | 0.02 |
ENST00000420223.1 ENST00000383768.2 |
ZCWPW2 |
zinc finger, CW type with PWWP domain 2 |
| chr5_-_9630463 | 0.02 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr17_-_33446735 | 0.02 |
ENST00000460118.2 ENST00000335858.7 |
RAD51D |
RAD51 paralog D |
| chr16_+_6069072 | 0.02 |
ENST00000547605.1 ENST00000550418.1 ENST00000553186.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_+_44502597 | 0.02 |
ENST00000260649.6 ENST00000409387.1 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr16_-_66764119 | 0.01 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr20_+_31805131 | 0.01 |
ENST00000375454.3 ENST00000375452.3 |
BPIFA3 |
BPI fold containing family A, member 3 |
| chr5_+_174151536 | 0.01 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr1_+_28199047 | 0.01 |
ENST00000373925.1 ENST00000328928.7 ENST00000373927.3 ENST00000427466.1 ENST00000442118.1 ENST00000373921.3 |
THEMIS2 |
thymocyte selection associated family member 2 |
| chr17_-_64187973 | 0.01 |
ENST00000583358.1 ENST00000392769.2 |
CEP112 |
centrosomal protein 112kDa |
| chr4_-_169239921 | 0.01 |
ENST00000514995.1 ENST00000393743.3 |
DDX60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr11_+_46958248 | 0.01 |
ENST00000536126.1 ENST00000278460.7 ENST00000378618.2 ENST00000395460.2 ENST00000378615.3 ENST00000543718.1 |
C11orf49 |
chromosome 11 open reading frame 49 |
| chr5_+_55149150 | 0.01 |
ENST00000297015.3 |
IL31RA |
interleukin 31 receptor A |
| chr11_-_111649015 | 0.01 |
ENST00000529841.1 |
RP11-108O10.2 |
RP11-108O10.2 |
| chr4_-_109541539 | 0.01 |
ENST00000509984.1 ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1 |
RPL34 antisense RNA 1 (head to head) |
| chr12_+_54674482 | 0.01 |
ENST00000547708.1 ENST00000340913.6 ENST00000551702.1 ENST00000330752.8 ENST00000547276.1 |
HNRNPA1 |
heterogeneous nuclear ribonucleoprotein A1 |
| chr18_+_22040593 | 0.01 |
ENST00000256906.4 |
HRH4 |
histamine receptor H4 |
| chr7_+_55433131 | 0.01 |
ENST00000254770.2 |
LANCL2 |
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr22_-_36903069 | 0.01 |
ENST00000216187.6 ENST00000423980.1 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr3_+_50273625 | 0.01 |
ENST00000536647.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr18_+_22040620 | 0.01 |
ENST00000426880.2 |
HRH4 |
histamine receptor H4 |
| chr17_-_64225508 | 0.01 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr5_+_140579162 | 0.01 |
ENST00000536699.1 ENST00000354757.3 |
PCDHB11 |
protocadherin beta 11 |
| chr19_-_19302931 | 0.01 |
ENST00000444486.3 ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B MEF2BNB MEF2B |
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr5_+_135394840 | 0.01 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr12_-_10282836 | 0.01 |
ENST00000304084.8 ENST00000353231.5 ENST00000525605.1 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr15_+_40697988 | 0.01 |
ENST00000487418.2 ENST00000479013.2 |
IVD |
isovaleryl-CoA dehydrogenase |
| chr8_-_62602327 | 0.01 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr13_+_78315295 | 0.01 |
ENST00000351546.3 |
SLAIN1 |
SLAIN motif family, member 1 |
| chr1_-_150669500 | 0.01 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr2_-_89442621 | 0.01 |
ENST00000492167.1 |
IGKV3-20 |
immunoglobulin kappa variable 3-20 |
| chr17_-_39203519 | 0.01 |
ENST00000542137.1 ENST00000391419.3 |
KRTAP2-1 |
keratin associated protein 2-1 |
| chr19_+_21324827 | 0.01 |
ENST00000600692.1 ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431 |
zinc finger protein 431 |
| chr14_+_57671888 | 0.01 |
ENST00000391612.1 |
AL391152.1 |
AL391152.1 |
| chr7_-_81399438 | 0.01 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr22_-_36903101 | 0.01 |
ENST00000397224.4 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr11_+_92085262 | 0.01 |
ENST00000298047.6 ENST00000409404.2 ENST00000541502.1 |
FAT3 |
FAT atypical cadherin 3 |
| chr17_-_39341594 | 0.01 |
ENST00000398472.1 |
KRTAP4-1 |
keratin associated protein 4-1 |
| chr3_+_62304648 | 0.01 |
ENST00000462069.1 ENST00000232519.5 ENST00000465142.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr9_-_132383055 | 0.01 |
ENST00000372478.4 |
C9orf50 |
chromosome 9 open reading frame 50 |
| chr14_+_105864847 | 0.01 |
ENST00000451127.2 |
TEX22 |
testis expressed 22 |
| chr1_+_84630645 | 0.01 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_+_43238438 | 0.01 |
ENST00000593138.1 ENST00000586681.1 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
| chr17_+_28443799 | 0.01 |
ENST00000584423.1 ENST00000247026.5 |
NSRP1 |
nuclear speckle splicing regulatory protein 1 |
| chr1_-_68698197 | 0.01 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chrX_-_18690210 | 0.01 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
| chr3_+_97887544 | 0.01 |
ENST00000356526.2 |
OR5H15 |
olfactory receptor, family 5, subfamily H, member 15 |
| chr5_-_55412774 | 0.01 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr9_+_95709733 | 0.01 |
ENST00000375482.3 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
| chr17_-_39254391 | 0.01 |
ENST00000333822.4 |
KRTAP4-8 |
keratin associated protein 4-8 |
| chrX_+_107334895 | 0.01 |
ENST00000372232.3 ENST00000345734.3 ENST00000372254.3 |
ATG4A |
autophagy related 4A, cysteine peptidase |
| chr12_-_71031220 | 0.01 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr16_+_72088376 | 0.01 |
ENST00000570083.1 ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP HPR |
haptoglobin haptoglobin-related protein |
| chr1_+_44679113 | 0.01 |
ENST00000361745.6 ENST00000446292.1 ENST00000440641.1 ENST00000436069.1 ENST00000437511.1 |
DMAP1 |
DNA methyltransferase 1 associated protein 1 |
| chr4_+_159727272 | 0.01 |
ENST00000379346.3 |
FNIP2 |
folliculin interacting protein 2 |
| chr5_+_129083772 | 0.01 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr10_+_99205959 | 0.01 |
ENST00000352634.4 ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr6_+_26402465 | 0.01 |
ENST00000476549.2 ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr14_-_71609965 | 0.01 |
ENST00000602957.1 |
RP6-91H8.3 |
RP6-91H8.3 |
| chr10_+_99205894 | 0.01 |
ENST00000370854.3 ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr2_-_68052694 | 0.01 |
ENST00000457448.1 |
AC010987.6 |
AC010987.6 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.0 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.0 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |