Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for ZIC3_ZIC4
Z-value: 0.41
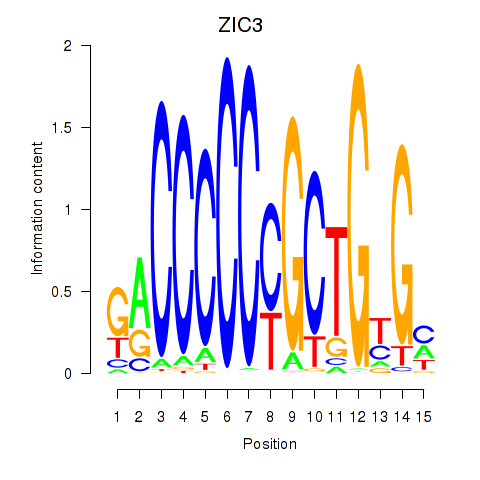
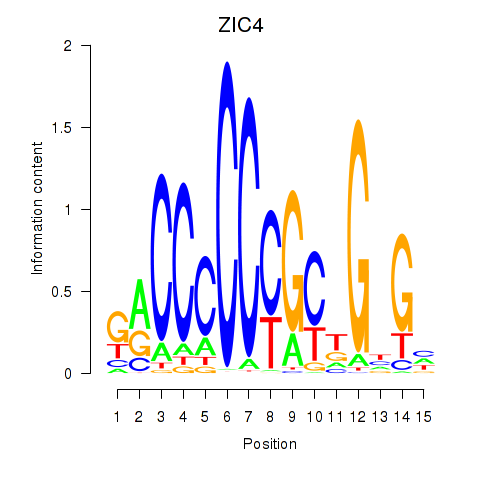
Transcription factors associated with ZIC3_ZIC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC3
|
ENSG00000156925.7 | ZIC3 |
|
ZIC4
|
ENSG00000174963.13 | ZIC4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC4 | hg19_v2_chr3_-_147124547_147124647 | 0.46 | 2.5e-01 | Click! |
| ZIC3 | hg19_v2_chrX_+_136648297_136648319 | 0.19 | 6.5e-01 | Click! |
Activity profile of ZIC3_ZIC4 motif
Sorted Z-values of ZIC3_ZIC4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC3_ZIC4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_75699149 | 1.31 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr5_+_75699040 | 1.21 |
ENST00000274364.6 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr17_-_43045439 | 0.59 |
ENST00000253407.3 |
C1QL1 |
complement component 1, q subcomponent-like 1 |
| chrX_+_51927919 | 0.54 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr12_+_56075330 | 0.42 |
ENST00000394252.3 |
METTL7B |
methyltransferase like 7B |
| chr14_-_30396802 | 0.31 |
ENST00000415220.2 |
PRKD1 |
protein kinase D1 |
| chr2_-_99552620 | 0.31 |
ENST00000428096.1 ENST00000397899.2 ENST00000420294.1 |
KIAA1211L |
KIAA1211-like |
| chr2_+_128180842 | 0.31 |
ENST00000402125.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr7_+_149571045 | 0.29 |
ENST00000479613.1 ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
| chr10_+_13142075 | 0.28 |
ENST00000378757.2 ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN |
optineurin |
| chr10_+_13142225 | 0.28 |
ENST00000378747.3 |
OPTN |
optineurin |
| chr2_+_8822113 | 0.28 |
ENST00000396290.1 ENST00000331129.3 |
ID2 |
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr21_+_37507210 | 0.25 |
ENST00000290354.5 |
CBR3 |
carbonyl reductase 3 |
| chr16_+_4364762 | 0.25 |
ENST00000262366.3 |
GLIS2 |
GLIS family zinc finger 2 |
| chr4_+_52917451 | 0.23 |
ENST00000295213.4 ENST00000419395.2 |
SPATA18 |
spermatogenesis associated 18 |
| chr16_+_30996502 | 0.23 |
ENST00000353250.5 ENST00000262520.6 ENST00000297679.5 ENST00000562932.1 ENST00000574447.1 |
HSD3B7 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr5_+_126626498 | 0.23 |
ENST00000503335.2 ENST00000508365.1 ENST00000418761.2 ENST00000274473.6 |
MEGF10 |
multiple EGF-like-domains 10 |
| chr3_+_54156570 | 0.22 |
ENST00000415676.2 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr17_+_38599693 | 0.22 |
ENST00000542955.1 ENST00000269593.4 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
| chr5_-_126366500 | 0.22 |
ENST00000308660.5 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
| chr3_+_54156664 | 0.22 |
ENST00000474759.1 ENST00000288197.5 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr3_+_12330560 | 0.22 |
ENST00000397026.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr14_+_100848311 | 0.22 |
ENST00000542471.2 |
WDR25 |
WD repeat domain 25 |
| chr11_-_2906979 | 0.21 |
ENST00000380725.1 ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C |
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr19_-_58609570 | 0.21 |
ENST00000600845.1 ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18 |
zinc finger and SCAN domain containing 18 |
| chr10_+_13141441 | 0.21 |
ENST00000263036.5 |
OPTN |
optineurin |
| chr3_-_112356944 | 0.21 |
ENST00000461431.1 |
CCDC80 |
coiled-coil domain containing 80 |
| chr1_+_38273818 | 0.20 |
ENST00000373042.4 |
C1orf122 |
chromosome 1 open reading frame 122 |
| chr1_+_38273988 | 0.20 |
ENST00000446260.2 |
C1orf122 |
chromosome 1 open reading frame 122 |
| chr20_+_34203794 | 0.20 |
ENST00000374273.3 |
SPAG4 |
sperm associated antigen 4 |
| chr17_+_78194205 | 0.19 |
ENST00000573809.1 ENST00000361193.3 ENST00000574967.1 ENST00000576126.1 ENST00000411502.3 ENST00000546047.2 |
SLC26A11 |
solute carrier family 26 (anion exchanger), member 11 |
| chr10_+_13141585 | 0.19 |
ENST00000378764.2 |
OPTN |
optineurin |
| chr2_-_45236540 | 0.19 |
ENST00000303077.6 |
SIX2 |
SIX homeobox 2 |
| chr4_+_74718906 | 0.18 |
ENST00000226524.3 |
PF4V1 |
platelet factor 4 variant 1 |
| chr1_+_215256467 | 0.18 |
ENST00000391894.2 ENST00000444842.2 |
KCNK2 |
potassium channel, subfamily K, member 2 |
| chr2_+_23608064 | 0.18 |
ENST00000486442.1 |
KLHL29 |
kelch-like family member 29 |
| chr11_+_64781575 | 0.17 |
ENST00000246747.4 ENST00000529384.1 |
ARL2 |
ADP-ribosylation factor-like 2 |
| chr19_-_40791211 | 0.17 |
ENST00000579047.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr9_+_4490394 | 0.17 |
ENST00000262352.3 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr2_+_20646824 | 0.17 |
ENST00000272233.4 |
RHOB |
ras homolog family member B |
| chr12_+_51785057 | 0.17 |
ENST00000535225.2 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr14_+_24630465 | 0.17 |
ENST00000557894.1 ENST00000559284.1 ENST00000560275.1 |
IRF9 |
interferon regulatory factor 9 |
| chr19_+_45409011 | 0.16 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr16_+_2039946 | 0.16 |
ENST00000248121.2 ENST00000568896.1 |
SYNGR3 |
synaptogyrin 3 |
| chr11_+_64781657 | 0.15 |
ENST00000533729.1 |
ARL2 |
ADP-ribosylation factor-like 2 |
| chr13_+_35516390 | 0.15 |
ENST00000540320.1 ENST00000400445.3 ENST00000310336.4 |
NBEA |
neurobeachin |
| chr17_-_39191107 | 0.15 |
ENST00000344363.5 |
KRTAP1-3 |
keratin associated protein 1-3 |
| chr5_-_151304337 | 0.15 |
ENST00000455880.2 ENST00000545569.1 ENST00000274576.4 |
GLRA1 |
glycine receptor, alpha 1 |
| chr19_+_49617581 | 0.14 |
ENST00000391864.3 |
LIN7B |
lin-7 homolog B (C. elegans) |
| chr1_+_43148625 | 0.14 |
ENST00000436427.1 |
YBX1 |
Y box binding protein 1 |
| chr5_-_14871866 | 0.14 |
ENST00000284268.6 |
ANKH |
ANKH inorganic pyrophosphate transport regulator |
| chr5_+_140753444 | 0.14 |
ENST00000517434.1 |
PCDHGA6 |
protocadherin gamma subfamily A, 6 |
| chr11_-_67272794 | 0.14 |
ENST00000436757.2 ENST00000356404.3 |
PITPNM1 |
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr19_-_51587502 | 0.14 |
ENST00000156499.2 ENST00000391802.1 |
KLK14 |
kallikrein-related peptidase 14 |
| chr1_+_52521797 | 0.13 |
ENST00000313334.8 |
BTF3L4 |
basic transcription factor 3-like 4 |
| chr17_-_39183452 | 0.13 |
ENST00000361883.5 |
KRTAP1-5 |
keratin associated protein 1-5 |
| chr5_-_111091948 | 0.13 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr10_-_79397479 | 0.13 |
ENST00000404771.3 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr4_+_38869298 | 0.13 |
ENST00000510213.1 ENST00000515037.1 |
FAM114A1 |
family with sequence similarity 114, member A1 |
| chr1_+_201617264 | 0.13 |
ENST00000367296.4 |
NAV1 |
neuron navigator 1 |
| chr17_-_78194147 | 0.13 |
ENST00000534910.1 ENST00000326317.6 |
SGSH |
N-sulfoglucosamine sulfohydrolase |
| chr6_-_85473156 | 0.13 |
ENST00000606784.1 ENST00000606325.1 |
TBX18 |
T-box 18 |
| chr1_+_201617450 | 0.13 |
ENST00000295624.6 ENST00000367297.4 ENST00000367300.3 |
NAV1 |
neuron navigator 1 |
| chr19_+_49617609 | 0.13 |
ENST00000221459.2 ENST00000486217.2 |
LIN7B |
lin-7 homolog B (C. elegans) |
| chrX_+_153686614 | 0.13 |
ENST00000369682.3 |
PLXNA3 |
plexin A3 |
| chr1_+_33352036 | 0.12 |
ENST00000373467.3 |
HPCA |
hippocalcin |
| chr1_+_52521957 | 0.12 |
ENST00000472944.2 ENST00000484036.1 |
BTF3L4 |
basic transcription factor 3-like 4 |
| chr11_-_65150103 | 0.12 |
ENST00000294187.6 ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45 |
solute carrier family 25, member 45 |
| chr6_+_89791507 | 0.12 |
ENST00000354922.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr2_+_105050794 | 0.12 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr11_-_74109422 | 0.12 |
ENST00000298198.4 |
PGM2L1 |
phosphoglucomutase 2-like 1 |
| chrX_-_34675391 | 0.11 |
ENST00000275954.3 |
TMEM47 |
transmembrane protein 47 |
| chr17_+_39394250 | 0.11 |
ENST00000254072.6 |
KRTAP9-8 |
keratin associated protein 9-8 |
| chr7_+_106809406 | 0.11 |
ENST00000468410.1 ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1 |
HMG-box transcription factor 1 |
| chr22_-_41636929 | 0.11 |
ENST00000216241.9 |
CHADL |
chondroadherin-like |
| chr10_+_35416223 | 0.11 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr19_+_13135790 | 0.11 |
ENST00000358552.3 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_-_56150184 | 0.11 |
ENST00000394554.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr17_-_7531121 | 0.10 |
ENST00000573566.1 ENST00000269298.5 |
SAT2 |
spermidine/spermine N1-acetyltransferase family member 2 |
| chr2_-_10220538 | 0.10 |
ENST00000381813.4 |
CYS1 |
cystin 1 |
| chr2_+_48757278 | 0.10 |
ENST00000404752.1 ENST00000406226.1 |
STON1 |
stonin 1 |
| chr1_+_52521928 | 0.10 |
ENST00000489308.2 |
BTF3L4 |
basic transcription factor 3-like 4 |
| chr6_+_33168637 | 0.10 |
ENST00000374677.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr6_+_33168597 | 0.10 |
ENST00000374675.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr2_+_37571845 | 0.10 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr4_+_38869410 | 0.09 |
ENST00000358869.2 |
FAM114A1 |
family with sequence similarity 114, member A1 |
| chr8_-_27462822 | 0.09 |
ENST00000522098.1 |
CLU |
clusterin |
| chr19_+_45251804 | 0.09 |
ENST00000164227.5 |
BCL3 |
B-cell CLL/lymphoma 3 |
| chr21_+_38445539 | 0.09 |
ENST00000418766.1 ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3 |
tetratricopeptide repeat domain 3 |
| chr2_-_201374781 | 0.09 |
ENST00000359878.3 ENST00000409157.1 |
KCTD18 |
potassium channel tetramerization domain containing 18 |
| chr3_+_11196206 | 0.09 |
ENST00000431010.2 |
HRH1 |
histamine receptor H1 |
| chr3_-_9811595 | 0.09 |
ENST00000256460.3 |
CAMK1 |
calcium/calmodulin-dependent protein kinase I |
| chr20_+_49411523 | 0.09 |
ENST00000371608.2 |
BCAS4 |
breast carcinoma amplified sequence 4 |
| chr5_+_140718396 | 0.09 |
ENST00000394576.2 |
PCDHGA2 |
protocadherin gamma subfamily A, 2 |
| chr2_+_37571717 | 0.09 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr20_-_31172598 | 0.09 |
ENST00000201961.2 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr17_+_38219063 | 0.09 |
ENST00000584985.1 ENST00000264637.4 ENST00000450525.2 |
THRA |
thyroid hormone receptor, alpha |
| chr10_-_79397202 | 0.09 |
ENST00000372437.1 ENST00000372408.2 ENST00000372403.4 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr5_-_131563501 | 0.09 |
ENST00000401867.1 ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr17_+_260097 | 0.09 |
ENST00000360127.6 ENST00000571106.1 ENST00000491373.1 |
C17orf97 |
chromosome 17 open reading frame 97 |
| chr14_-_23822080 | 0.09 |
ENST00000397267.1 ENST00000354772.3 |
SLC22A17 |
solute carrier family 22, member 17 |
| chr5_-_76383133 | 0.09 |
ENST00000255198.2 |
ZBED3 |
zinc finger, BED-type containing 3 |
| chr1_-_43232649 | 0.08 |
ENST00000372526.2 ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1 |
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr19_-_50143452 | 0.08 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr2_+_101436564 | 0.08 |
ENST00000335681.5 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr5_-_176923803 | 0.08 |
ENST00000506161.1 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
| chr1_-_154928562 | 0.08 |
ENST00000368463.3 ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1 |
pre-B-cell leukemia homeobox interacting protein 1 |
| chr5_+_139027877 | 0.08 |
ENST00000302517.3 |
CXXC5 |
CXXC finger protein 5 |
| chr1_+_43148059 | 0.08 |
ENST00000321358.7 ENST00000332220.6 |
YBX1 |
Y box binding protein 1 |
| chr5_-_176923846 | 0.08 |
ENST00000506537.1 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
| chr17_-_73178599 | 0.08 |
ENST00000578238.1 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr1_+_156024552 | 0.08 |
ENST00000368304.5 ENST00000368302.3 |
LAMTOR2 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr12_+_108523489 | 0.08 |
ENST00000261400.3 |
WSCD2 |
WSC domain containing 2 |
| chr10_-_88854518 | 0.08 |
ENST00000277865.4 |
GLUD1 |
glutamate dehydrogenase 1 |
| chr4_+_81951957 | 0.08 |
ENST00000282701.2 |
BMP3 |
bone morphogenetic protein 3 |
| chr19_+_54926601 | 0.08 |
ENST00000301194.4 |
TTYH1 |
tweety family member 1 |
| chr19_+_33685490 | 0.08 |
ENST00000253193.7 |
LRP3 |
low density lipoprotein receptor-related protein 3 |
| chr4_+_115519577 | 0.08 |
ENST00000310836.6 |
UGT8 |
UDP glycosyltransferase 8 |
| chr12_+_108523133 | 0.07 |
ENST00000547525.1 |
WSCD2 |
WSC domain containing 2 |
| chr5_-_178157700 | 0.07 |
ENST00000335815.2 |
ZNF354A |
zinc finger protein 354A |
| chr16_+_2198604 | 0.07 |
ENST00000210187.6 |
RAB26 |
RAB26, member RAS oncogene family |
| chr13_-_52378231 | 0.07 |
ENST00000280056.2 ENST00000444610.2 |
DHRS12 |
dehydrogenase/reductase (SDR family) member 12 |
| chr17_-_19619917 | 0.07 |
ENST00000325411.5 ENST00000350657.5 ENST00000433844.2 |
SLC47A2 |
solute carrier family 47 (multidrug and toxin extrusion), member 2 |
| chr17_-_1083078 | 0.07 |
ENST00000574266.1 ENST00000302538.5 |
ABR |
active BCR-related |
| chrX_+_102631844 | 0.07 |
ENST00000372634.1 ENST00000299872.7 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr19_-_48752812 | 0.07 |
ENST00000359009.4 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr10_+_85954377 | 0.07 |
ENST00000332904.3 ENST00000372117.3 |
CDHR1 |
cadherin-related family member 1 |
| chr20_+_3869423 | 0.07 |
ENST00000497424.1 |
PANK2 |
pantothenate kinase 2 |
| chr11_-_2160180 | 0.07 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr19_-_48753104 | 0.07 |
ENST00000447740.2 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr11_-_73694346 | 0.07 |
ENST00000310473.3 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr3_+_195413160 | 0.07 |
ENST00000599448.1 |
LINC00969 |
long intergenic non-protein coding RNA 969 |
| chr2_+_238768187 | 0.07 |
ENST00000254661.4 ENST00000409726.1 |
RAMP1 |
receptor (G protein-coupled) activity modifying protein 1 |
| chr1_+_156024525 | 0.07 |
ENST00000368305.4 |
LAMTOR2 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr9_-_98279241 | 0.07 |
ENST00000437951.1 ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1 |
patched 1 |
| chr11_-_2158507 | 0.07 |
ENST00000381392.1 ENST00000381395.1 ENST00000418738.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr10_+_134145735 | 0.07 |
ENST00000368613.4 |
LRRC27 |
leucine rich repeat containing 27 |
| chr1_+_43232913 | 0.07 |
ENST00000372525.5 ENST00000536543.1 |
C1orf50 |
chromosome 1 open reading frame 50 |
| chr10_+_28966271 | 0.07 |
ENST00000375533.3 |
BAMBI |
BMP and activin membrane-bound inhibitor |
| chr1_+_6845578 | 0.07 |
ENST00000467404.2 ENST00000439411.2 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr20_-_56286479 | 0.07 |
ENST00000265626.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr8_-_27850141 | 0.07 |
ENST00000524352.1 |
SCARA5 |
scavenger receptor class A, member 5 (putative) |
| chr13_+_27131798 | 0.07 |
ENST00000361042.4 |
WASF3 |
WAS protein family, member 3 |
| chr1_-_146644036 | 0.07 |
ENST00000425272.2 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr10_+_45455207 | 0.07 |
ENST00000334940.6 ENST00000374417.2 ENST00000340258.5 ENST00000427758.1 |
RASSF4 |
Ras association (RalGDS/AF-6) domain family member 4 |
| chr12_-_57824739 | 0.07 |
ENST00000347140.3 ENST00000402412.1 |
R3HDM2 |
R3H domain containing 2 |
| chr22_-_47134077 | 0.07 |
ENST00000541677.1 ENST00000216264.8 |
CERK |
ceramide kinase |
| chrX_+_102631248 | 0.07 |
ENST00000361298.4 ENST00000372645.3 ENST00000372635.1 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr1_-_212873267 | 0.07 |
ENST00000243440.1 |
BATF3 |
basic leucine zipper transcription factor, ATF-like 3 |
| chr5_-_176924562 | 0.07 |
ENST00000359895.2 ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
| chr21_-_45196326 | 0.07 |
ENST00000291568.5 |
CSTB |
cystatin B (stefin B) |
| chr20_+_3870024 | 0.06 |
ENST00000610179.1 |
PANK2 |
pantothenate kinase 2 |
| chr16_-_1993260 | 0.06 |
ENST00000361871.3 |
MSRB1 |
methionine sulfoxide reductase B1 |
| chr19_+_54926621 | 0.06 |
ENST00000376530.3 ENST00000445095.1 ENST00000391739.3 ENST00000376531.3 |
TTYH1 |
tweety family member 1 |
| chr7_+_37888199 | 0.06 |
ENST00000199447.4 ENST00000455500.1 |
NME8 |
NME/NM23 family member 8 |
| chr11_-_67275542 | 0.06 |
ENST00000531506.1 |
CDK2AP2 |
cyclin-dependent kinase 2 associated protein 2 |
| chr17_-_42276574 | 0.06 |
ENST00000589805.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr9_-_130742792 | 0.06 |
ENST00000373095.1 |
FAM102A |
family with sequence similarity 102, member A |
| chr11_-_32457075 | 0.06 |
ENST00000448076.3 |
WT1 |
Wilms tumor 1 |
| chr14_+_105781048 | 0.06 |
ENST00000458164.2 ENST00000447393.1 |
PACS2 |
phosphofurin acidic cluster sorting protein 2 |
| chr17_+_45000483 | 0.06 |
ENST00000576910.2 ENST00000439730.2 ENST00000393456.2 ENST00000415811.2 ENST00000575949.1 ENST00000225567.4 ENST00000572403.1 ENST00000570879.1 |
GOSR2 |
golgi SNAP receptor complex member 2 |
| chr6_-_38607673 | 0.06 |
ENST00000481247.1 |
BTBD9 |
BTB (POZ) domain containing 9 |
| chr7_+_37888555 | 0.06 |
ENST00000444718.1 ENST00000440017.1 |
NME8 |
NME/NM23 family member 8 |
| chr13_-_99404875 | 0.06 |
ENST00000376503.5 |
SLC15A1 |
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr2_-_106015527 | 0.06 |
ENST00000344213.4 ENST00000358129.4 |
FHL2 |
four and a half LIM domains 2 |
| chr1_+_85527987 | 0.06 |
ENST00000326813.8 ENST00000294664.6 ENST00000528899.1 |
WDR63 |
WD repeat domain 63 |
| chr19_-_18717627 | 0.06 |
ENST00000392386.3 |
CRLF1 |
cytokine receptor-like factor 1 |
| chr5_+_76506706 | 0.06 |
ENST00000340978.3 ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B |
phosphodiesterase 8B |
| chr12_-_59314246 | 0.06 |
ENST00000320743.3 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr3_-_17783990 | 0.06 |
ENST00000429383.4 ENST00000446863.1 ENST00000414349.1 ENST00000428355.1 ENST00000425944.1 ENST00000445294.1 ENST00000444471.1 ENST00000415814.2 |
TBC1D5 |
TBC1 domain family, member 5 |
| chr19_+_37096194 | 0.06 |
ENST00000460670.1 ENST00000292928.2 ENST00000439428.1 |
ZNF382 |
zinc finger protein 382 |
| chr1_-_182641037 | 0.06 |
ENST00000483095.2 |
RGS8 |
regulator of G-protein signaling 8 |
| chr1_-_182640988 | 0.06 |
ENST00000367556.1 |
RGS8 |
regulator of G-protein signaling 8 |
| chr12_-_121734489 | 0.06 |
ENST00000412367.2 ENST00000402834.4 ENST00000404169.3 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr16_-_18470696 | 0.06 |
ENST00000427999.2 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr16_-_31085514 | 0.06 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr17_-_73179046 | 0.06 |
ENST00000314523.7 ENST00000420826.2 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr15_-_90294523 | 0.05 |
ENST00000300057.4 |
MESP1 |
mesoderm posterior 1 homolog (mouse) |
| chr5_+_132202252 | 0.05 |
ENST00000378670.3 ENST00000378667.1 ENST00000378665.1 |
UQCRQ |
ubiquinol-cytochrome c reductase, complex III subunit VII, 9.5kDa |
| chr22_-_19165917 | 0.05 |
ENST00000451283.1 |
SLC25A1 |
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr3_+_50712672 | 0.05 |
ENST00000266037.9 |
DOCK3 |
dedicator of cytokinesis 3 |
| chr1_+_218519577 | 0.05 |
ENST00000366930.4 ENST00000366929.4 |
TGFB2 |
transforming growth factor, beta 2 |
| chr20_-_3154162 | 0.05 |
ENST00000360342.3 |
LZTS3 |
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr19_+_13906250 | 0.05 |
ENST00000254323.2 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
| chr21_+_45285050 | 0.05 |
ENST00000291572.8 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr16_-_8962853 | 0.05 |
ENST00000565287.1 ENST00000311052.5 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
| chr19_-_53193731 | 0.05 |
ENST00000598536.1 ENST00000594682.2 ENST00000601257.1 |
ZNF83 |
zinc finger protein 83 |
| chr1_+_114471809 | 0.05 |
ENST00000426820.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr3_-_99833333 | 0.05 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr22_+_23134974 | 0.05 |
ENST00000390314.2 |
IGLV2-11 |
immunoglobulin lambda variable 2-11 |
| chr11_-_119211525 | 0.05 |
ENST00000528368.1 |
C1QTNF5 |
C1q and tumor necrosis factor related protein 5 |
| chr12_+_111471828 | 0.05 |
ENST00000261726.6 |
CUX2 |
cut-like homeobox 2 |
| chr17_-_15903002 | 0.05 |
ENST00000399277.1 |
ZSWIM7 |
zinc finger, SWIM-type containing 7 |
| chr7_+_66205712 | 0.05 |
ENST00000451741.2 ENST00000442563.1 ENST00000450873.2 ENST00000284957.5 |
KCTD7 RABGEF1 |
potassium channel tetramerization domain containing 7 RAB guanine nucleotide exchange factor (GEF) 1 |
| chr2_+_109150850 | 0.05 |
ENST00000544547.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr1_-_31381528 | 0.05 |
ENST00000339394.6 |
SDC3 |
syndecan 3 |
| chr15_-_66790146 | 0.05 |
ENST00000316634.5 |
SNAPC5 |
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr13_+_27131887 | 0.05 |
ENST00000335327.5 |
WASF3 |
WAS protein family, member 3 |
| chr4_-_15656923 | 0.05 |
ENST00000382358.4 ENST00000412094.2 ENST00000341285.3 |
FBXL5 |
F-box and leucine-rich repeat protein 5 |
| chr17_+_39261584 | 0.05 |
ENST00000391415.1 |
KRTAP4-9 |
keratin associated protein 4-9 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 2.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.3 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.3 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.2 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.0 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.1 | GO:0071484 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.1 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 | 0.1 | GO:0003129 | heart induction(GO:0003129) regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.0 | GO:0097212 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) regulation of hydrogen peroxide catabolic process(GO:2000295) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 2.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |