Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
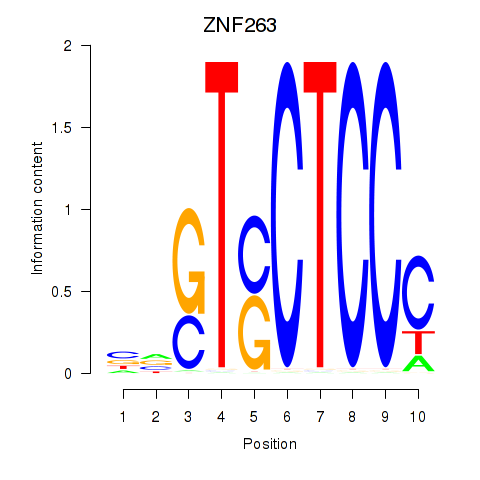
Results for ZNF263
Z-value: 0.19
Transcription factors associated with ZNF263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF263
|
ENSG00000006194.6 | ZNF263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF263 | hg19_v2_chr16_+_3313791_3313834 | 0.55 | 1.6e-01 | Click! |
Activity profile of ZNF263 motif
Sorted Z-values of ZNF263 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF263
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_55658687 | 0.34 |
ENST00000593046.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr6_+_150464155 | 0.30 |
ENST00000361131.4 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr19_-_55658281 | 0.25 |
ENST00000585321.2 ENST00000587465.2 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr14_-_21994525 | 0.22 |
ENST00000538754.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr19_-_55658650 | 0.21 |
ENST00000589226.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr19_+_55587266 | 0.17 |
ENST00000201647.6 ENST00000540810.1 |
EPS8L1 |
EPS8-like 1 |
| chr19_-_51504852 | 0.16 |
ENST00000391806.2 ENST00000347619.4 ENST00000291726.7 ENST00000320838.5 |
KLK8 |
kallikrein-related peptidase 8 |
| chr19_-_51456198 | 0.16 |
ENST00000594846.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr15_-_90358048 | 0.16 |
ENST00000300060.6 ENST00000560137.1 |
ANPEP |
alanyl (membrane) aminopeptidase |
| chr16_-_11370330 | 0.15 |
ENST00000241808.4 ENST00000435245.2 |
PRM2 |
protamine 2 |
| chr22_+_29876197 | 0.15 |
ENST00000310624.6 |
NEFH |
neurofilament, heavy polypeptide |
| chr12_+_4382917 | 0.14 |
ENST00000261254.3 |
CCND2 |
cyclin D2 |
| chr8_+_95653302 | 0.13 |
ENST00000423620.2 ENST00000433389.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr18_+_61420169 | 0.13 |
ENST00000425392.1 ENST00000336429.2 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr12_+_6930703 | 0.12 |
ENST00000311268.3 |
GPR162 |
G protein-coupled receptor 162 |
| chr11_-_46142505 | 0.12 |
ENST00000524497.1 ENST00000418153.2 |
PHF21A |
PHD finger protein 21A |
| chr6_-_32157947 | 0.12 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr1_-_153588765 | 0.12 |
ENST00000368701.1 ENST00000344616.2 |
S100A14 |
S100 calcium binding protein A14 |
| chr1_-_153588334 | 0.12 |
ENST00000476873.1 |
S100A14 |
S100 calcium binding protein A14 |
| chr8_+_95653373 | 0.12 |
ENST00000358397.5 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr12_+_6930813 | 0.12 |
ENST00000428545.2 |
GPR162 |
G protein-coupled receptor 162 |
| chr8_+_95653427 | 0.12 |
ENST00000454170.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr19_-_36001286 | 0.11 |
ENST00000602679.1 ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN |
dermokine |
| chr12_+_54943134 | 0.11 |
ENST00000243052.3 |
PDE1B |
phosphodiesterase 1B, calmodulin-dependent |
| chr12_+_6930964 | 0.11 |
ENST00000382315.3 |
GPR162 |
G protein-coupled receptor 162 |
| chr2_+_89890533 | 0.11 |
ENST00000429992.2 |
IGKV2D-40 |
immunoglobulin kappa variable 2D-40 |
| chr20_-_22565101 | 0.11 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr14_+_75746340 | 0.11 |
ENST00000555686.1 ENST00000555672.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr19_-_49258606 | 0.11 |
ENST00000310160.3 |
FUT1 |
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| chr6_-_136847610 | 0.10 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr1_-_6321035 | 0.10 |
ENST00000377893.2 |
GPR153 |
G protein-coupled receptor 153 |
| chr1_+_60280458 | 0.10 |
ENST00000455990.1 ENST00000371208.3 |
HOOK1 |
hook microtubule-tethering protein 1 |
| chr12_-_25055949 | 0.10 |
ENST00000539282.1 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr19_+_54371114 | 0.10 |
ENST00000448420.1 ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM |
myeloid-associated differentiation marker |
| chr6_+_36097992 | 0.10 |
ENST00000211287.4 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chr1_+_55505184 | 0.10 |
ENST00000302118.5 |
PCSK9 |
proprotein convertase subtilisin/kexin type 9 |
| chr6_+_36098262 | 0.09 |
ENST00000373761.6 ENST00000373766.5 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chr3_+_189507432 | 0.09 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr15_-_63674034 | 0.09 |
ENST00000344366.3 ENST00000422263.2 |
CA12 |
carbonic anhydrase XII |
| chr2_+_103236004 | 0.09 |
ENST00000233969.2 |
SLC9A2 |
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr14_-_77608121 | 0.09 |
ENST00000319374.4 |
ZDHHC22 |
zinc finger, DHHC-type containing 22 |
| chr3_-_46735155 | 0.09 |
ENST00000318962.4 |
ALS2CL |
ALS2 C-terminal like |
| chr1_+_211432700 | 0.09 |
ENST00000452621.2 |
RCOR3 |
REST corepressor 3 |
| chr19_-_49565254 | 0.09 |
ENST00000593537.1 |
NTF4 |
neurotrophin 4 |
| chr12_-_54779511 | 0.09 |
ENST00000551109.1 ENST00000546970.1 |
ZNF385A |
zinc finger protein 385A |
| chr7_-_44265492 | 0.09 |
ENST00000425809.1 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr1_+_35220613 | 0.09 |
ENST00000338513.1 |
GJB5 |
gap junction protein, beta 5, 31.1kDa |
| chr19_-_51487071 | 0.08 |
ENST00000391807.1 ENST00000593904.1 |
KLK7 |
kallikrein-related peptidase 7 |
| chr7_-_139876812 | 0.08 |
ENST00000397560.2 |
JHDM1D |
lysine (K)-specific demethylase 7A |
| chr2_-_31361543 | 0.08 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_+_205473720 | 0.08 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr17_-_8027402 | 0.08 |
ENST00000541682.2 ENST00000317814.4 ENST00000577735.1 |
HES7 |
hes family bHLH transcription factor 7 |
| chr17_-_39674668 | 0.08 |
ENST00000393981.3 |
KRT15 |
keratin 15 |
| chr10_-_90342947 | 0.08 |
ENST00000437752.1 ENST00000331772.4 |
RNLS |
renalase, FAD-dependent amine oxidase |
| chr19_+_44037334 | 0.08 |
ENST00000314228.5 |
ZNF575 |
zinc finger protein 575 |
| chr16_+_86544113 | 0.08 |
ENST00000262426.4 |
FOXF1 |
forkhead box F1 |
| chr3_-_13921594 | 0.07 |
ENST00000285018.4 |
WNT7A |
wingless-type MMTV integration site family, member 7A |
| chr9_+_35673853 | 0.07 |
ENST00000378357.4 |
CA9 |
carbonic anhydrase IX |
| chrX_-_129402857 | 0.07 |
ENST00000447817.1 ENST00000370978.4 |
ZNF280C |
zinc finger protein 280C |
| chr18_+_33877654 | 0.07 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr6_+_69345166 | 0.07 |
ENST00000370598.1 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr9_-_124991124 | 0.07 |
ENST00000394319.4 ENST00000340587.3 |
LHX6 |
LIM homeobox 6 |
| chr17_+_35294075 | 0.07 |
ENST00000254457.5 |
LHX1 |
LIM homeobox 1 |
| chr15_+_91427642 | 0.07 |
ENST00000328850.3 ENST00000414248.2 |
FES |
feline sarcoma oncogene |
| chr5_+_167718604 | 0.07 |
ENST00000265293.4 |
WWC1 |
WW and C2 domain containing 1 |
| chr2_-_238499337 | 0.07 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr19_+_44037546 | 0.07 |
ENST00000601282.1 |
ZNF575 |
zinc finger protein 575 |
| chr6_-_41130841 | 0.07 |
ENST00000373122.4 |
TREM2 |
triggering receptor expressed on myeloid cells 2 |
| chr8_+_124194875 | 0.07 |
ENST00000522648.1 ENST00000276699.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr6_+_18155632 | 0.07 |
ENST00000297792.5 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr12_-_6798523 | 0.07 |
ENST00000319770.3 |
ZNF384 |
zinc finger protein 384 |
| chr8_+_31497271 | 0.07 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr6_+_18155560 | 0.07 |
ENST00000546309.2 ENST00000388870.2 ENST00000397244.1 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr1_+_167599330 | 0.07 |
ENST00000367854.3 ENST00000361496.3 |
RCSD1 |
RCSD domain containing 1 |
| chr22_+_18593446 | 0.06 |
ENST00000316027.6 |
TUBA8 |
tubulin, alpha 8 |
| chr14_-_94856987 | 0.06 |
ENST00000449399.3 ENST00000404814.4 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr6_+_90272027 | 0.06 |
ENST00000522441.1 |
ANKRD6 |
ankyrin repeat domain 6 |
| chr7_-_98030360 | 0.06 |
ENST00000005260.8 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
| chr11_+_1860682 | 0.06 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr12_-_6798616 | 0.06 |
ENST00000355772.4 ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384 |
zinc finger protein 384 |
| chr1_+_171107241 | 0.06 |
ENST00000236166.3 |
FMO6P |
flavin containing monooxygenase 6 pseudogene |
| chr6_-_41130914 | 0.06 |
ENST00000373113.3 ENST00000338469.3 |
TREM2 |
triggering receptor expressed on myeloid cells 2 |
| chr6_+_154360553 | 0.06 |
ENST00000452687.2 |
OPRM1 |
opioid receptor, mu 1 |
| chr17_-_42200958 | 0.06 |
ENST00000336057.5 |
HDAC5 |
histone deacetylase 5 |
| chr12_-_6798410 | 0.06 |
ENST00000361959.3 ENST00000436774.2 ENST00000544482.1 |
ZNF384 |
zinc finger protein 384 |
| chr17_+_77020146 | 0.06 |
ENST00000579760.1 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr6_+_154360616 | 0.06 |
ENST00000229768.5 ENST00000419506.2 ENST00000524163.1 ENST00000414028.2 ENST00000435918.2 ENST00000337049.4 |
OPRM1 |
opioid receptor, mu 1 |
| chr8_+_124194752 | 0.06 |
ENST00000318462.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr16_+_57662138 | 0.06 |
ENST00000562414.1 ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr19_-_44306590 | 0.06 |
ENST00000377950.3 |
LYPD5 |
LY6/PLAUR domain containing 5 |
| chr6_+_41514078 | 0.06 |
ENST00000373063.3 ENST00000373060.1 |
FOXP4 |
forkhead box P4 |
| chr6_+_154360476 | 0.06 |
ENST00000428397.2 |
OPRM1 |
opioid receptor, mu 1 |
| chr17_-_50236039 | 0.06 |
ENST00000451037.2 |
CA10 |
carbonic anhydrase X |
| chr1_+_212782012 | 0.06 |
ENST00000341491.4 ENST00000366985.1 |
ATF3 |
activating transcription factor 3 |
| chr4_+_53728457 | 0.06 |
ENST00000248706.3 |
RASL11B |
RAS-like, family 11, member B |
| chr16_+_57662419 | 0.06 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr17_-_42200996 | 0.06 |
ENST00000587135.1 ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5 |
histone deacetylase 5 |
| chr12_-_54785074 | 0.06 |
ENST00000338010.5 ENST00000550774.1 |
ZNF385A |
zinc finger protein 385A |
| chr10_+_119301928 | 0.06 |
ENST00000553456.3 |
EMX2 |
empty spiracles homeobox 2 |
| chr2_+_191045656 | 0.06 |
ENST00000443551.2 |
C2orf88 |
chromosome 2 open reading frame 88 |
| chr6_-_13487784 | 0.06 |
ENST00000379287.3 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chrX_-_117107542 | 0.06 |
ENST00000371878.1 |
KLHL13 |
kelch-like family member 13 |
| chr20_-_18038521 | 0.06 |
ENST00000278780.6 |
OVOL2 |
ovo-like zinc finger 2 |
| chr12_+_53773944 | 0.06 |
ENST00000551969.1 ENST00000327443.4 |
SP1 |
Sp1 transcription factor |
| chr11_+_1860200 | 0.06 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr1_+_15250596 | 0.06 |
ENST00000361144.5 |
KAZN |
kazrin, periplakin interacting protein |
| chr11_-_101000445 | 0.06 |
ENST00000534013.1 |
PGR |
progesterone receptor |
| chrX_-_21776281 | 0.06 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr2_+_18059906 | 0.06 |
ENST00000304101.4 |
KCNS3 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr10_+_50822480 | 0.06 |
ENST00000455728.2 |
CHAT |
choline O-acetyltransferase |
| chr12_-_25055177 | 0.06 |
ENST00000538118.1 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr18_-_47721447 | 0.06 |
ENST00000285039.7 |
MYO5B |
myosin VB |
| chr12_-_53625958 | 0.06 |
ENST00000327550.3 ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG |
retinoic acid receptor, gamma |
| chr11_-_72492903 | 0.06 |
ENST00000537947.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr19_-_51143075 | 0.06 |
ENST00000600079.1 ENST00000593901.1 |
SYT3 |
synaptotagmin III |
| chr19_-_47975417 | 0.06 |
ENST00000236877.6 |
SLC8A2 |
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr14_+_23775971 | 0.06 |
ENST00000250405.5 |
BCL2L2 |
BCL2-like 2 |
| chr19_-_54984354 | 0.06 |
ENST00000301200.2 |
CDC42EP5 |
CDC42 effector protein (Rho GTPase binding) 5 |
| chr11_-_72492878 | 0.06 |
ENST00000535054.1 ENST00000545082.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr14_+_93799556 | 0.06 |
ENST00000256339.4 |
UNC79 |
unc-79 homolog (C. elegans) |
| chr4_-_185395672 | 0.06 |
ENST00000393593.3 |
IRF2 |
interferon regulatory factor 2 |
| chrX_-_117107680 | 0.06 |
ENST00000447671.2 ENST00000262820.3 |
KLHL13 |
kelch-like family member 13 |
| chr17_-_42452063 | 0.06 |
ENST00000588098.1 |
ITGA2B |
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr10_+_23983671 | 0.05 |
ENST00000376462.1 |
KIAA1217 |
KIAA1217 |
| chr4_-_84256024 | 0.05 |
ENST00000311412.5 |
HPSE |
heparanase |
| chr15_-_63674218 | 0.05 |
ENST00000178638.3 |
CA12 |
carbonic anhydrase XII |
| chr15_+_74218787 | 0.05 |
ENST00000261921.7 |
LOXL1 |
lysyl oxidase-like 1 |
| chr1_+_156119466 | 0.05 |
ENST00000414683.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr12_-_54785054 | 0.05 |
ENST00000352268.6 ENST00000549962.1 |
ZNF385A |
zinc finger protein 385A |
| chr21_-_27945562 | 0.05 |
ENST00000299340.4 ENST00000435845.2 |
CYYR1 |
cysteine/tyrosine-rich 1 |
| chr20_+_45523227 | 0.05 |
ENST00000327619.5 ENST00000357410.3 |
EYA2 |
eyes absent homolog 2 (Drosophila) |
| chr19_+_917287 | 0.05 |
ENST00000592648.1 ENST00000234371.5 |
KISS1R |
KISS1 receptor |
| chr11_+_7597639 | 0.05 |
ENST00000533792.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr2_+_39893043 | 0.05 |
ENST00000281961.2 |
TMEM178A |
transmembrane protein 178A |
| chr16_+_56995854 | 0.05 |
ENST00000566128.1 |
CETP |
cholesteryl ester transfer protein, plasma |
| chr1_-_159915386 | 0.05 |
ENST00000361509.3 ENST00000368094.1 |
IGSF9 |
immunoglobulin superfamily, member 9 |
| chr11_+_56949221 | 0.05 |
ENST00000497933.1 |
LRRC55 |
leucine rich repeat containing 55 |
| chr14_-_65409438 | 0.05 |
ENST00000557049.1 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr1_+_209859510 | 0.05 |
ENST00000367028.2 ENST00000261465.1 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr6_+_30848557 | 0.05 |
ENST00000460944.2 ENST00000324771.8 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chrX_+_107069063 | 0.05 |
ENST00000262843.6 |
MID2 |
midline 2 |
| chr8_-_75233563 | 0.05 |
ENST00000342232.4 |
JPH1 |
junctophilin 1 |
| chr3_-_46735132 | 0.05 |
ENST00000415953.1 |
ALS2CL |
ALS2 C-terminal like |
| chr14_-_21566731 | 0.05 |
ENST00000360947.3 |
ZNF219 |
zinc finger protein 219 |
| chr14_-_65409502 | 0.05 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr11_-_130184470 | 0.05 |
ENST00000357899.4 ENST00000397753.1 |
ZBTB44 |
zinc finger and BTB domain containing 44 |
| chrX_-_24665208 | 0.05 |
ENST00000356768.4 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr15_-_68497657 | 0.05 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr5_-_88179302 | 0.05 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr4_-_153457197 | 0.05 |
ENST00000281708.4 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_-_90759440 | 0.05 |
ENST00000336904.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr9_-_116061145 | 0.05 |
ENST00000416588.2 |
RNF183 |
ring finger protein 183 |
| chr6_-_53409890 | 0.05 |
ENST00000229416.6 |
GCLC |
glutamate-cysteine ligase, catalytic subunit |
| chr2_-_218867711 | 0.05 |
ENST00000446903.1 |
TNS1 |
tensin 1 |
| chr11_-_72853091 | 0.05 |
ENST00000311172.7 ENST00000409314.1 |
FCHSD2 |
FCH and double SH3 domains 2 |
| chr19_-_49864746 | 0.05 |
ENST00000598810.1 |
TEAD2 |
TEA domain family member 2 |
| chr18_-_74207146 | 0.05 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr4_+_7194247 | 0.05 |
ENST00000507866.2 |
SORCS2 |
sortilin-related VPS10 domain containing receptor 2 |
| chr7_-_150038704 | 0.05 |
ENST00000466675.1 ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2 |
retinoic acid receptor responder (tazarotene induced) 2 |
| chr18_-_52626622 | 0.05 |
ENST00000591504.1 |
CCDC68 |
coiled-coil domain containing 68 |
| chr3_+_189507523 | 0.05 |
ENST00000437221.1 ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63 |
tumor protein p63 |
| chr11_-_100999775 | 0.05 |
ENST00000263463.5 |
PGR |
progesterone receptor |
| chr22_+_38035459 | 0.05 |
ENST00000357436.4 |
SH3BP1 |
SH3-domain binding protein 1 |
| chr11_-_118023490 | 0.04 |
ENST00000324727.4 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
| chr16_+_30671223 | 0.04 |
ENST00000568722.1 |
FBRS |
fibrosin |
| chr1_-_204329013 | 0.04 |
ENST00000272203.3 ENST00000414478.1 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
| chrX_+_23352133 | 0.04 |
ENST00000379361.4 |
PTCHD1 |
patched domain containing 1 |
| chr10_-_25305011 | 0.04 |
ENST00000331161.4 ENST00000376363.1 |
ENKUR |
enkurin, TRPC channel interacting protein |
| chr16_-_28550320 | 0.04 |
ENST00000395641.2 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr15_+_85427903 | 0.04 |
ENST00000286749.3 ENST00000394573.1 ENST00000537703.1 |
SLC28A1 |
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr4_+_113970772 | 0.04 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr6_+_4776580 | 0.04 |
ENST00000397588.3 |
CDYL |
chromodomain protein, Y-like |
| chr1_+_211432775 | 0.04 |
ENST00000419091.2 |
RCOR3 |
REST corepressor 3 |
| chr2_-_71062938 | 0.04 |
ENST00000410009.3 |
CD207 |
CD207 molecule, langerin |
| chr6_+_7541845 | 0.04 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr17_-_4890649 | 0.04 |
ENST00000361571.5 |
CAMTA2 |
calmodulin binding transcription activator 2 |
| chr2_-_60780702 | 0.04 |
ENST00000359629.5 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr6_+_41606176 | 0.04 |
ENST00000441667.1 ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI |
MyoD family inhibitor |
| chr3_-_8693755 | 0.04 |
ENST00000341795.3 |
SSUH2 |
ssu-2 homolog (C. elegans) |
| chr4_-_84255935 | 0.04 |
ENST00000513463.1 |
HPSE |
heparanase |
| chr19_+_6772710 | 0.04 |
ENST00000304076.2 ENST00000602142.1 ENST00000596764.1 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
| chr11_-_130184555 | 0.04 |
ENST00000525842.1 |
ZBTB44 |
zinc finger and BTB domain containing 44 |
| chr2_-_60780536 | 0.04 |
ENST00000538214.1 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr19_+_51728316 | 0.04 |
ENST00000436584.2 ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33 |
CD33 molecule |
| chr7_-_3214287 | 0.04 |
ENST00000404626.3 |
AC091801.1 |
LOC392621; Uncharacterized protein |
| chr17_+_42248063 | 0.04 |
ENST00000293414.1 |
ASB16 |
ankyrin repeat and SOCS box containing 16 |
| chr15_+_91427691 | 0.04 |
ENST00000559355.1 ENST00000394302.1 |
FES |
feline sarcoma oncogene |
| chr19_-_52035044 | 0.04 |
ENST00000359982.4 ENST00000436458.1 ENST00000425629.3 ENST00000391797.3 ENST00000343300.4 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
| chr17_+_8924837 | 0.04 |
ENST00000173229.2 |
NTN1 |
netrin 1 |
| chr8_+_19796381 | 0.04 |
ENST00000524029.1 ENST00000522701.1 ENST00000311322.8 |
LPL |
lipoprotein lipase |
| chr3_-_171178157 | 0.04 |
ENST00000465393.1 ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK |
TRAF2 and NCK interacting kinase |
| chr10_+_123872483 | 0.04 |
ENST00000369001.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr11_+_59480899 | 0.04 |
ENST00000300150.7 |
STX3 |
syntaxin 3 |
| chr19_-_52034971 | 0.04 |
ENST00000346477.3 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
| chr20_-_1306351 | 0.04 |
ENST00000381812.1 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chr10_+_80027105 | 0.04 |
ENST00000461034.1 ENST00000476909.1 ENST00000459633.1 |
LINC00595 |
long intergenic non-protein coding RNA 595 |
| chr20_-_1306391 | 0.04 |
ENST00000339987.3 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chr17_-_50235423 | 0.04 |
ENST00000340813.6 |
CA10 |
carbonic anhydrase X |
| chr6_-_112408661 | 0.04 |
ENST00000368662.5 |
TUBE1 |
tubulin, epsilon 1 |
| chr9_+_84304628 | 0.04 |
ENST00000437181.1 |
RP11-154D17.1 |
RP11-154D17.1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.1 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0032499 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.0 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.2 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:1904800 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.2 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) right lung development(GO:0060458) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.0 | GO:0071505 | response to mycophenolic acid(GO:0071505) cellular response to mycophenolic acid(GO:0071506) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.0 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.0 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.0 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.0 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.0 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.0 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.0 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.0 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.0 | GO:0010652 | pulmonary valve formation(GO:0003193) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.0 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.0 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.0 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |