Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
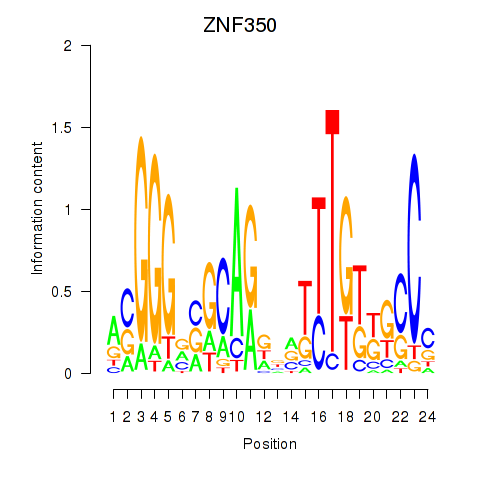
Results for ZNF350
Z-value: 0.24
Transcription factors associated with ZNF350
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF350
|
ENSG00000256683.2 | ZNF350 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF350 | hg19_v2_chr19_-_52489923_52490113 | -0.37 | 3.6e-01 | Click! |
Activity profile of ZNF350 motif
Sorted Z-values of ZNF350 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF350
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39674668 | 0.34 |
ENST00000393981.3 |
KRT15 |
keratin 15 |
| chr14_+_96671016 | 0.16 |
ENST00000542454.2 ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2 RP11-404P21.8 |
bradykinin receptor B2 Uncharacterized protein |
| chr10_+_50822480 | 0.16 |
ENST00000455728.2 |
CHAT |
choline O-acetyltransferase |
| chr11_-_112034803 | 0.13 |
ENST00000528832.1 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
| chr11_-_112034780 | 0.13 |
ENST00000524595.1 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
| chr11_-_112034831 | 0.13 |
ENST00000280357.7 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
| chr2_-_70780770 | 0.12 |
ENST00000444975.1 ENST00000445399.1 ENST00000418333.2 |
TGFA |
transforming growth factor, alpha |
| chr2_-_70780572 | 0.12 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr12_+_113376157 | 0.11 |
ENST00000228928.7 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr9_+_12693336 | 0.07 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr1_+_156119798 | 0.07 |
ENST00000355014.2 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr10_+_50822083 | 0.06 |
ENST00000337653.2 ENST00000395562.2 |
CHAT |
choline O-acetyltransferase |
| chr11_-_69633792 | 0.06 |
ENST00000334134.2 |
FGF3 |
fibroblast growth factor 3 |
| chr1_-_200379129 | 0.06 |
ENST00000367353.1 |
ZNF281 |
zinc finger protein 281 |
| chr11_+_118826999 | 0.05 |
ENST00000264031.2 |
UPK2 |
uroplakin 2 |
| chr4_+_120133791 | 0.05 |
ENST00000274030.6 |
USP53 |
ubiquitin specific peptidase 53 |
| chr1_-_200379180 | 0.05 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr1_-_46089718 | 0.05 |
ENST00000421127.2 ENST00000343901.2 ENST00000528266.1 |
CCDC17 |
coiled-coil domain containing 17 |
| chr9_+_124329336 | 0.05 |
ENST00000394340.3 ENST00000436835.1 ENST00000259371.2 |
DAB2IP |
DAB2 interacting protein |
| chr1_-_204654481 | 0.05 |
ENST00000367176.3 |
LRRN2 |
leucine rich repeat neuronal 2 |
| chr11_-_9113093 | 0.04 |
ENST00000450649.2 |
SCUBE2 |
signal peptide, CUB domain, EGF-like 2 |
| chr1_-_200379104 | 0.04 |
ENST00000367352.3 |
ZNF281 |
zinc finger protein 281 |
| chr12_-_93323013 | 0.04 |
ENST00000322349.8 |
EEA1 |
early endosome antigen 1 |
| chr12_+_50451462 | 0.04 |
ENST00000447966.2 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chr2_-_9143786 | 0.04 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr21_-_30365136 | 0.04 |
ENST00000361371.5 ENST00000389194.2 ENST00000389195.2 |
LTN1 |
listerin E3 ubiquitin protein ligase 1 |
| chr12_+_113376249 | 0.04 |
ENST00000551007.1 ENST00000548514.1 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr20_-_54580523 | 0.04 |
ENST00000064571.2 |
CBLN4 |
cerebellin 4 precursor |
| chr13_-_96329048 | 0.04 |
ENST00000606011.1 ENST00000499499.2 |
DNAJC3-AS1 |
DNAJC3 antisense RNA 1 (head to head) |
| chr8_-_119124045 | 0.04 |
ENST00000378204.2 |
EXT1 |
exostosin glycosyltransferase 1 |
| chr8_+_144295067 | 0.04 |
ENST00000330824.2 |
GPIHBP1 |
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr4_+_75858290 | 0.04 |
ENST00000513238.1 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr13_+_96329381 | 0.04 |
ENST00000602402.1 ENST00000376795.6 |
DNAJC3 |
DnaJ (Hsp40) homolog, subfamily C, member 3 |
| chr20_+_4666882 | 0.04 |
ENST00000379440.4 ENST00000430350.2 |
PRNP |
prion protein |
| chr2_-_106810783 | 0.03 |
ENST00000283148.7 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
| chr4_+_75858318 | 0.03 |
ENST00000307428.7 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr12_+_50451331 | 0.03 |
ENST00000228468.4 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chr16_-_74734672 | 0.03 |
ENST00000306247.7 ENST00000575686.1 |
MLKL |
mixed lineage kinase domain-like |
| chr17_-_74449252 | 0.03 |
ENST00000319380.7 |
UBE2O |
ubiquitin-conjugating enzyme E2O |
| chr2_-_106810742 | 0.03 |
ENST00000409501.3 ENST00000428048.2 ENST00000441952.1 ENST00000457835.1 ENST00000540130.1 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
| chr1_+_63788730 | 0.03 |
ENST00000371116.2 |
FOXD3 |
forkhead box D3 |
| chr20_+_4667094 | 0.03 |
ENST00000424424.1 ENST00000457586.1 |
PRNP |
prion protein |
| chr2_-_219157250 | 0.03 |
ENST00000434015.2 ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
| chr19_+_35645618 | 0.03 |
ENST00000392218.2 ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
| chr3_-_155524049 | 0.03 |
ENST00000534941.1 ENST00000340171.2 |
C3orf33 |
chromosome 3 open reading frame 33 |
| chr2_+_177025619 | 0.03 |
ENST00000410016.1 |
HOXD3 |
homeobox D3 |
| chr10_-_15762124 | 0.03 |
ENST00000378076.3 |
ITGA8 |
integrin, alpha 8 |
| chr16_+_2479390 | 0.03 |
ENST00000397066.4 |
CCNF |
cyclin F |
| chr11_-_9113137 | 0.03 |
ENST00000520467.1 ENST00000309263.3 ENST00000457346.2 |
SCUBE2 |
signal peptide, CUB domain, EGF-like 2 |
| chr17_-_39597173 | 0.03 |
ENST00000246646.3 |
KRT38 |
keratin 38 |
| chr15_+_42694573 | 0.03 |
ENST00000397200.4 ENST00000569827.1 |
CAPN3 |
calpain 3, (p94) |
| chr12_+_81471816 | 0.03 |
ENST00000261206.3 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
| chr1_-_247094628 | 0.03 |
ENST00000366508.1 ENST00000326225.3 ENST00000391829.2 |
AHCTF1 |
AT hook containing transcription factor 1 |
| chr17_+_68071389 | 0.03 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr18_+_77155942 | 0.03 |
ENST00000397790.2 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr16_-_74734742 | 0.03 |
ENST00000308807.7 ENST00000573267.1 |
MLKL |
mixed lineage kinase domain-like |
| chr4_-_76008706 | 0.03 |
ENST00000562355.1 ENST00000563602.1 |
RP11-44F21.5 |
RP11-44F21.5 |
| chr1_-_46089639 | 0.02 |
ENST00000445048.2 |
CCDC17 |
coiled-coil domain containing 17 |
| chr10_-_12084770 | 0.02 |
ENST00000357604.5 |
UPF2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr3_+_113775576 | 0.02 |
ENST00000485050.1 ENST00000281273.4 |
QTRTD1 |
queuine tRNA-ribosyltransferase domain containing 1 |
| chr19_+_35645817 | 0.02 |
ENST00000423817.3 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
| chr16_+_4526341 | 0.02 |
ENST00000458134.3 ENST00000219700.6 ENST00000575120.1 ENST00000572812.1 ENST00000574466.1 ENST00000576827.1 ENST00000570445.1 |
HMOX2 |
heme oxygenase (decycling) 2 |
| chr1_+_156611704 | 0.02 |
ENST00000329117.5 |
BCAN |
brevican |
| chr16_+_67282853 | 0.02 |
ENST00000299798.11 |
SLC9A5 |
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr15_-_65477637 | 0.02 |
ENST00000300107.3 |
CLPX |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
| chr17_+_68071458 | 0.02 |
ENST00000589377.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr14_-_94254821 | 0.02 |
ENST00000393140.1 |
PRIMA1 |
proline rich membrane anchor 1 |
| chr22_+_26565440 | 0.02 |
ENST00000404234.3 ENST00000529632.2 ENST00000360929.3 ENST00000248933.6 ENST00000343706.4 |
SEZ6L |
seizure related 6 homolog (mouse)-like |
| chr3_-_128212016 | 0.02 |
ENST00000498200.1 ENST00000341105.2 |
GATA2 |
GATA binding protein 2 |
| chr18_+_77155856 | 0.02 |
ENST00000253506.5 ENST00000591814.1 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr11_+_7041677 | 0.02 |
ENST00000299481.4 |
NLRP14 |
NLR family, pyrin domain containing 14 |
| chr1_+_204797749 | 0.02 |
ENST00000367172.4 ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC |
neurofascin |
| chr2_+_238600788 | 0.02 |
ENST00000289175.6 ENST00000244815.5 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
| chr3_-_11623804 | 0.01 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr5_+_176514413 | 0.01 |
ENST00000513166.1 |
FGFR4 |
fibroblast growth factor receptor 4 |
| chr12_-_109219937 | 0.01 |
ENST00000546697.1 |
SSH1 |
slingshot protein phosphatase 1 |
| chr8_-_100905363 | 0.01 |
ENST00000524245.1 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr11_-_7041466 | 0.01 |
ENST00000536068.1 ENST00000278314.4 |
ZNF214 |
zinc finger protein 214 |
| chr3_-_128902759 | 0.01 |
ENST00000422453.2 ENST00000504813.1 ENST00000512338.1 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chrX_+_56259316 | 0.01 |
ENST00000468660.1 |
KLF8 |
Kruppel-like factor 8 |
| chr5_-_93077293 | 0.01 |
ENST00000510627.4 |
POU5F2 |
POU domain class 5, transcription factor 2 |
| chrX_-_120090454 | 0.01 |
ENST00000434883.2 |
CT47A7 |
cancer/testis antigen family 47, member A7 |
| chr11_+_48002076 | 0.01 |
ENST00000418331.2 ENST00000440289.2 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
| chr16_+_71879861 | 0.01 |
ENST00000427980.2 ENST00000568581.1 |
ATXN1L IST1 |
ataxin 1-like increased sodium tolerance 1 homolog (yeast) |
| chr17_-_39968855 | 0.01 |
ENST00000355468.3 ENST00000590496.1 |
LEPREL4 |
leprecan-like 4 |
| chr4_-_174255400 | 0.01 |
ENST00000506267.1 |
HMGB2 |
high mobility group box 2 |
| chr11_+_64323428 | 0.01 |
ENST00000377581.3 |
SLC22A11 |
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr1_+_156611960 | 0.01 |
ENST00000361588.5 |
BCAN |
brevican |
| chr19_+_5681011 | 0.01 |
ENST00000581893.1 ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L |
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr15_-_43029252 | 0.01 |
ENST00000563260.1 ENST00000356231.3 |
CDAN1 |
codanin 1 |
| chr17_+_48624450 | 0.01 |
ENST00000006658.6 ENST00000356488.4 ENST00000393244.3 |
SPATA20 |
spermatogenesis associated 20 |
| chr3_-_128902729 | 0.01 |
ENST00000451728.2 ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr18_+_45778672 | 0.01 |
ENST00000600091.1 |
AC091150.1 |
HCG1818186; Uncharacterized protein |
| chr3_+_50229037 | 0.01 |
ENST00000232461.3 ENST00000433068.1 |
GNAT1 |
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
| chr17_+_7487146 | 0.01 |
ENST00000396501.4 ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1 |
mannose-P-dolichol utilization defect 1 |
| chr2_-_157198860 | 0.01 |
ENST00000409572.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr4_-_174255536 | 0.01 |
ENST00000446922.2 |
HMGB2 |
high mobility group box 2 |
| chr1_+_22970119 | 0.01 |
ENST00000374640.4 ENST00000374639.3 ENST00000374637.1 |
C1QC |
complement component 1, q subcomponent, C chain |
| chr17_-_18950950 | 0.01 |
ENST00000284154.5 |
GRAP |
GRB2-related adaptor protein |
| chrX_-_47479246 | 0.00 |
ENST00000295987.7 ENST00000340666.4 |
SYN1 |
synapsin I |
| chr19_+_42829702 | 0.00 |
ENST00000334370.4 |
MEGF8 |
multiple EGF-like-domains 8 |
| chr19_+_1450112 | 0.00 |
ENST00000590469.1 ENST00000233607.2 ENST00000238483.4 ENST00000590877.1 |
APC2 |
adenomatosis polyposis coli 2 |
| chr1_+_228395755 | 0.00 |
ENST00000284548.11 ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN |
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr19_+_5681153 | 0.00 |
ENST00000579559.1 ENST00000577222.1 |
HSD11B1L RPL36 |
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr3_-_113775328 | 0.00 |
ENST00000483766.1 ENST00000545063.1 ENST00000491000.1 ENST00000295878.3 |
KIAA1407 |
KIAA1407 |
| chr11_+_68671310 | 0.00 |
ENST00000255078.3 ENST00000539224.1 |
IGHMBP2 |
immunoglobulin mu binding protein 2 |
| chrX_-_17878827 | 0.00 |
ENST00000360011.1 |
RAI2 |
retinoic acid induced 2 |
| chr14_+_73603126 | 0.00 |
ENST00000557356.1 ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1 |
presenilin 1 |
| chr17_-_79881408 | 0.00 |
ENST00000392366.3 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr20_+_44035847 | 0.00 |
ENST00000372712.2 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.0 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |