Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
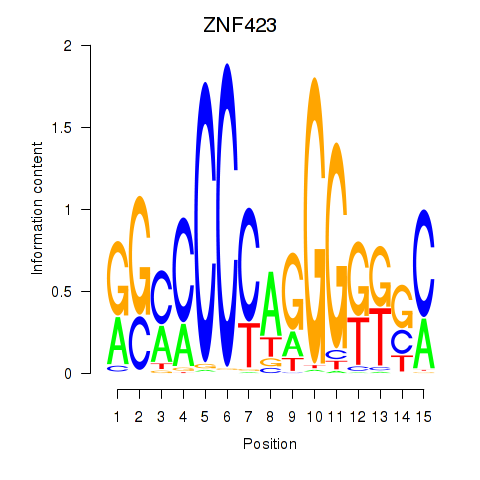
Results for ZNF423
Z-value: 0.25
Transcription factors associated with ZNF423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF423
|
ENSG00000102935.7 | ZNF423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF423 | hg19_v2_chr16_-_49890016_49890046 | -0.83 | 1.2e-02 | Click! |
Activity profile of ZNF423 motif
Sorted Z-values of ZNF423 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF423
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_44265492 | 0.35 |
ENST00000425809.1 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr8_-_42234745 | 0.17 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr12_+_7342441 | 0.13 |
ENST00000412720.2 ENST00000396637.3 |
PEX5 |
peroxisomal biogenesis factor 5 |
| chr12_+_7342271 | 0.13 |
ENST00000434354.2 ENST00000544456.1 ENST00000545574.1 ENST00000420616.2 |
PEX5 |
peroxisomal biogenesis factor 5 |
| chr2_-_46769694 | 0.12 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr17_+_7341586 | 0.11 |
ENST00000575235.1 |
FGF11 |
fibroblast growth factor 11 |
| chr16_+_3014217 | 0.11 |
ENST00000572045.1 |
KREMEN2 |
kringle containing transmembrane protein 2 |
| chr12_-_54779511 | 0.10 |
ENST00000551109.1 ENST00000546970.1 |
ZNF385A |
zinc finger protein 385A |
| chr1_-_33366931 | 0.10 |
ENST00000373463.3 ENST00000329151.5 |
TMEM54 |
transmembrane protein 54 |
| chr7_-_19184929 | 0.10 |
ENST00000275461.3 |
FERD3L |
Fer3-like bHLH transcription factor |
| chr11_-_44972390 | 0.10 |
ENST00000395648.3 ENST00000531928.2 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr10_+_85933494 | 0.10 |
ENST00000372126.3 |
C10orf99 |
chromosome 10 open reading frame 99 |
| chr11_-_44972418 | 0.10 |
ENST00000525680.1 ENST00000528290.1 ENST00000530035.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr19_-_11689752 | 0.09 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr11_+_60691924 | 0.09 |
ENST00000544065.1 ENST00000453848.2 ENST00000005286.4 |
TMEM132A |
transmembrane protein 132A |
| chr9_-_124976185 | 0.09 |
ENST00000464484.2 |
LHX6 |
LIM homeobox 6 |
| chr22_-_37823468 | 0.08 |
ENST00000402918.2 |
ELFN2 |
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr9_-_124976154 | 0.08 |
ENST00000482062.1 |
LHX6 |
LIM homeobox 6 |
| chr12_+_7342178 | 0.08 |
ENST00000266563.5 ENST00000543974.1 |
PEX5 |
peroxisomal biogenesis factor 5 |
| chr16_+_3014269 | 0.08 |
ENST00000575885.1 ENST00000571007.1 ENST00000319500.6 |
KREMEN2 |
kringle containing transmembrane protein 2 |
| chr5_-_176057518 | 0.07 |
ENST00000393693.2 |
SNCB |
synuclein, beta |
| chr7_+_20370300 | 0.07 |
ENST00000537992.1 |
ITGB8 |
integrin, beta 8 |
| chr5_-_176057365 | 0.07 |
ENST00000310112.3 |
SNCB |
synuclein, beta |
| chr1_-_48462566 | 0.07 |
ENST00000606738.2 |
TRABD2B |
TraB domain containing 2B |
| chr4_+_4388805 | 0.06 |
ENST00000504171.1 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr4_+_152330390 | 0.06 |
ENST00000503146.1 ENST00000435205.1 |
FAM160A1 |
family with sequence similarity 160, member A1 |
| chr11_-_44972476 | 0.06 |
ENST00000527685.1 ENST00000308212.5 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr14_-_77608121 | 0.06 |
ENST00000319374.4 |
ZDHHC22 |
zinc finger, DHHC-type containing 22 |
| chr11_-_44971702 | 0.06 |
ENST00000533940.1 ENST00000533937.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr11_-_47207950 | 0.06 |
ENST00000298838.6 ENST00000531226.1 ENST00000524509.1 ENST00000528201.1 ENST00000530513.1 |
PACSIN3 |
protein kinase C and casein kinase substrate in neurons 3 |
| chr16_-_11681023 | 0.06 |
ENST00000570904.1 ENST00000574701.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr14_-_93582148 | 0.05 |
ENST00000267615.6 ENST00000553452.1 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
| chr1_-_203055129 | 0.05 |
ENST00000241651.4 |
MYOG |
myogenin (myogenic factor 4) |
| chr12_-_57522813 | 0.05 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr8_-_80680078 | 0.05 |
ENST00000337919.5 ENST00000354724.3 |
HEY1 |
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr5_+_68788594 | 0.05 |
ENST00000396442.2 ENST00000380766.2 |
OCLN |
occludin |
| chr8_+_30241934 | 0.05 |
ENST00000538486.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr6_+_151186554 | 0.05 |
ENST00000367321.3 ENST00000367307.4 |
MTHFD1L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr14_-_93582214 | 0.05 |
ENST00000556603.2 ENST00000354313.3 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
| chr7_+_20370746 | 0.05 |
ENST00000222573.4 |
ITGB8 |
integrin, beta 8 |
| chr6_+_151187074 | 0.05 |
ENST00000367308.4 |
MTHFD1L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr10_-_76859247 | 0.05 |
ENST00000472493.2 ENST00000605915.1 ENST00000478873.2 |
DUSP13 |
dual specificity phosphatase 13 |
| chr16_+_27325202 | 0.04 |
ENST00000395762.2 ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R |
interleukin 4 receptor |
| chr5_+_170736243 | 0.04 |
ENST00000296921.5 |
TLX3 |
T-cell leukemia homeobox 3 |
| chr9_+_116638562 | 0.04 |
ENST00000374126.5 ENST00000288466.7 |
ZNF618 |
zinc finger protein 618 |
| chr16_-_11681316 | 0.04 |
ENST00000571688.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr11_-_61062762 | 0.04 |
ENST00000335613.5 |
VWCE |
von Willebrand factor C and EGF domains |
| chr2_+_12246664 | 0.04 |
ENST00000449986.1 |
AC096559.1 |
AC096559.1 |
| chr10_+_135340859 | 0.04 |
ENST00000252945.3 ENST00000421586.1 ENST00000418356.1 |
CYP2E1 |
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chrX_-_136113833 | 0.04 |
ENST00000298110.1 |
GPR101 |
G protein-coupled receptor 101 |
| chrX_-_48827976 | 0.04 |
ENST00000218176.3 |
KCND1 |
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr11_-_64013663 | 0.04 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr6_+_33589161 | 0.04 |
ENST00000605930.1 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr4_+_128982490 | 0.04 |
ENST00000394288.3 ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B |
La ribonucleoprotein domain family, member 1B |
| chr7_-_27219849 | 0.03 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr16_+_30675654 | 0.03 |
ENST00000287468.5 ENST00000395073.2 |
FBRS |
fibrosin |
| chr1_-_204121013 | 0.03 |
ENST00000367201.3 |
ETNK2 |
ethanolamine kinase 2 |
| chr6_-_143266297 | 0.03 |
ENST00000367603.2 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
| chr9_-_35619539 | 0.03 |
ENST00000396757.1 |
CD72 |
CD72 molecule |
| chr9_-_131486367 | 0.03 |
ENST00000372663.4 ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12 |
zinc finger, DHHC-type containing 12 |
| chr3_-_195808952 | 0.03 |
ENST00000540528.1 ENST00000392396.3 ENST00000535031.1 ENST00000420415.1 |
TFRC |
transferrin receptor |
| chr6_-_32160622 | 0.03 |
ENST00000487761.1 ENST00000375040.3 |
GPSM3 |
G-protein signaling modulator 3 |
| chr1_-_204121102 | 0.03 |
ENST00000367202.4 |
ETNK2 |
ethanolamine kinase 2 |
| chr4_-_120548779 | 0.03 |
ENST00000264805.5 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
| chr22_+_35653445 | 0.03 |
ENST00000420166.1 ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4 |
HMG box domain containing 4 |
| chr17_-_48227877 | 0.03 |
ENST00000316878.6 |
PPP1R9B |
protein phosphatase 1, regulatory subunit 9B |
| chr19_+_41770269 | 0.03 |
ENST00000378215.4 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr1_-_204121298 | 0.03 |
ENST00000367199.2 |
ETNK2 |
ethanolamine kinase 2 |
| chr17_+_36584662 | 0.03 |
ENST00000431231.2 ENST00000437668.3 |
ARHGAP23 |
Rho GTPase activating protein 23 |
| chr19_+_41770349 | 0.03 |
ENST00000602130.1 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr21_-_18985158 | 0.03 |
ENST00000339775.6 |
BTG3 |
BTG family, member 3 |
| chr12_-_50101003 | 0.03 |
ENST00000550488.1 |
FMNL3 |
formin-like 3 |
| chr19_-_57183114 | 0.03 |
ENST00000537055.2 ENST00000601659.1 |
ZNF835 |
zinc finger protein 835 |
| chr2_+_135011731 | 0.03 |
ENST00000281923.2 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr3_-_195808980 | 0.03 |
ENST00000360110.4 |
TFRC |
transferrin receptor |
| chr19_+_41770236 | 0.02 |
ENST00000392006.3 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr21_-_18985230 | 0.02 |
ENST00000457956.1 ENST00000348354.6 |
BTG3 |
BTG family, member 3 |
| chr19_-_4066890 | 0.02 |
ENST00000322357.4 |
ZBTB7A |
zinc finger and BTB domain containing 7A |
| chr6_-_91296737 | 0.02 |
ENST00000369332.3 ENST00000369329.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr18_-_60985914 | 0.02 |
ENST00000589955.1 |
BCL2 |
B-cell CLL/lymphoma 2 |
| chr2_+_62932779 | 0.02 |
ENST00000427809.1 ENST00000405482.1 ENST00000431489.1 |
EHBP1 |
EH domain binding protein 1 |
| chr12_+_52643077 | 0.02 |
ENST00000553310.2 ENST00000544024.1 |
KRT86 |
keratin 86 |
| chr17_-_73178599 | 0.02 |
ENST00000578238.1 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr4_-_166034029 | 0.02 |
ENST00000306480.6 |
TMEM192 |
transmembrane protein 192 |
| chr3_-_69129501 | 0.02 |
ENST00000540295.1 ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3 |
ubiquitin-like modifier activating enzyme 3 |
| chr10_+_124914285 | 0.02 |
ENST00000407911.2 |
BUB3 |
BUB3 mitotic checkpoint protein |
| chrX_+_106163626 | 0.02 |
ENST00000336803.1 |
CLDN2 |
claudin 2 |
| chr7_+_150748288 | 0.02 |
ENST00000490540.1 |
ASIC3 |
acid-sensing (proton-gated) ion channel 3 |
| chr6_-_91296602 | 0.02 |
ENST00000369325.3 ENST00000369327.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr4_+_128982430 | 0.02 |
ENST00000512292.1 ENST00000508819.1 |
LARP1B |
La ribonucleoprotein domain family, member 1B |
| chr4_+_4388245 | 0.02 |
ENST00000433139.2 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr6_-_35480640 | 0.02 |
ENST00000428978.1 ENST00000322263.4 |
TULP1 |
tubby like protein 1 |
| chr12_+_7037461 | 0.02 |
ENST00000396684.2 |
ATN1 |
atrophin 1 |
| chr8_+_30241995 | 0.02 |
ENST00000397323.4 ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS |
RNA binding protein with multiple splicing |
| chr9_-_130742792 | 0.02 |
ENST00000373095.1 |
FAM102A |
family with sequence similarity 102, member A |
| chr3_+_6902794 | 0.02 |
ENST00000357716.4 ENST00000486284.1 ENST00000389336.4 ENST00000403881.1 ENST00000402647.2 |
GRM7 |
glutamate receptor, metabotropic 7 |
| chr9_-_99382065 | 0.02 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chrX_-_1572629 | 0.02 |
ENST00000534940.1 |
ASMTL |
acetylserotonin O-methyltransferase-like |
| chr1_-_155177677 | 0.02 |
ENST00000368378.3 ENST00000541990.1 ENST00000457183.2 |
THBS3 |
thrombospondin 3 |
| chr19_+_10531150 | 0.02 |
ENST00000352831.6 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr6_-_35480705 | 0.02 |
ENST00000229771.6 |
TULP1 |
tubby like protein 1 |
| chr12_-_125348448 | 0.01 |
ENST00000339570.5 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr4_-_78740511 | 0.01 |
ENST00000504123.1 ENST00000264903.4 ENST00000515441.1 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
| chr12_-_118810688 | 0.01 |
ENST00000542532.1 ENST00000392533.3 |
TAOK3 |
TAO kinase 3 |
| chr13_-_46626847 | 0.01 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr20_-_25062767 | 0.01 |
ENST00000429762.3 ENST00000444511.2 ENST00000376707.3 |
VSX1 |
visual system homeobox 1 |
| chr1_+_53527854 | 0.01 |
ENST00000371500.3 ENST00000395871.2 ENST00000312553.5 |
PODN |
podocan |
| chr19_-_23941680 | 0.01 |
ENST00000402377.3 |
ZNF681 |
zinc finger protein 681 |
| chr12_-_4554780 | 0.01 |
ENST00000228837.2 |
FGF6 |
fibroblast growth factor 6 |
| chr4_+_4387983 | 0.01 |
ENST00000397958.1 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr6_+_30130969 | 0.01 |
ENST00000376694.4 |
TRIM15 |
tripartite motif containing 15 |
| chr19_+_45582453 | 0.01 |
ENST00000591607.1 ENST00000591747.1 ENST00000270257.4 ENST00000391951.2 ENST00000587566.1 |
GEMIN7 MARK4 |
gem (nuclear organelle) associated protein 7 MAP/microtubule affinity-regulating kinase 4 |
| chr10_+_124913793 | 0.01 |
ENST00000368865.4 ENST00000538238.1 ENST00000368859.2 |
BUB3 |
BUB3 mitotic checkpoint protein |
| chr1_-_35658736 | 0.01 |
ENST00000357214.5 |
SFPQ |
splicing factor proline/glutamine-rich |
| chr12_-_125348329 | 0.01 |
ENST00000546215.1 ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr21_-_45682099 | 0.01 |
ENST00000270172.3 ENST00000418993.1 |
DNMT3L |
DNA (cytosine-5-)-methyltransferase 3-like |
| chr20_+_23016057 | 0.01 |
ENST00000255008.3 |
SSTR4 |
somatostatin receptor 4 |
| chr7_-_30029367 | 0.01 |
ENST00000242059.5 |
SCRN1 |
secernin 1 |
| chr10_-_118032979 | 0.01 |
ENST00000355422.6 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr10_-_121296045 | 0.01 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr6_+_142468361 | 0.01 |
ENST00000367630.4 |
VTA1 |
vesicle (multivesicular body) trafficking 1 |
| chr3_+_180630444 | 0.01 |
ENST00000491062.1 ENST00000468861.1 ENST00000445140.2 ENST00000484958.1 |
FXR1 |
fragile X mental retardation, autosomal homolog 1 |
| chr6_+_42896865 | 0.01 |
ENST00000372836.4 ENST00000394142.3 |
CNPY3 |
canopy FGF signaling regulator 3 |
| chr2_-_206950781 | 0.01 |
ENST00000403263.1 |
INO80D |
INO80 complex subunit D |
| chr6_+_30131318 | 0.01 |
ENST00000376688.1 |
TRIM15 |
tripartite motif containing 15 |
| chr11_-_6495101 | 0.00 |
ENST00000528227.1 ENST00000359518.3 ENST00000345851.3 ENST00000537602.1 |
TRIM3 |
tripartite motif containing 3 |
| chr22_+_39853258 | 0.00 |
ENST00000341184.6 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr11_-_6495082 | 0.00 |
ENST00000536344.1 |
TRIM3 |
tripartite motif containing 3 |
| chr2_+_171785824 | 0.00 |
ENST00000452526.2 |
GORASP2 |
golgi reassembly stacking protein 2, 55kDa |
| chr3_+_196439170 | 0.00 |
ENST00000392391.3 ENST00000314118.4 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr17_-_73179046 | 0.00 |
ENST00000314523.7 ENST00000420826.2 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr8_+_22423219 | 0.00 |
ENST00000523965.1 ENST00000521554.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr1_+_55271736 | 0.00 |
ENST00000358193.3 ENST00000371273.3 |
C1orf177 |
chromosome 1 open reading frame 177 |
| chr3_+_196439275 | 0.00 |
ENST00000296333.5 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr19_-_56110859 | 0.00 |
ENST00000221665.3 ENST00000592585.1 |
FIZ1 |
FLT3-interacting zinc finger 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901090 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.4 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:1903862 | regulation of muscle atrophy(GO:0014735) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0098845 | postsynaptic endosome(GO:0098845) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0052831 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |