Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for ZNF8
Z-value: 0.20
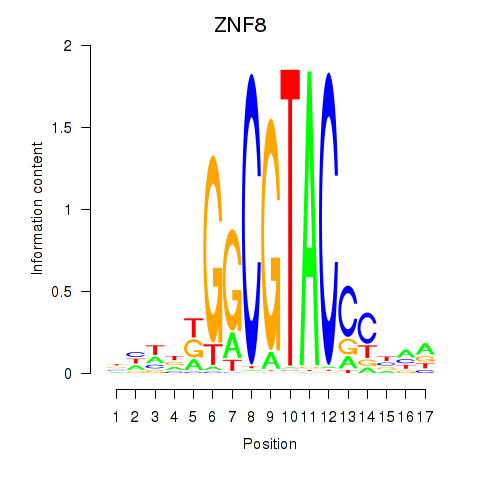
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000083842.8 | ZNF8 |
|
ZNF8
|
ENSG00000273439.1 | ZNF8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF8 | hg19_v2_chr19_+_58790314_58790358 | -0.68 | 6.5e-02 | Click! |
Activity profile of ZNF8 motif
Sorted Z-values of ZNF8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_35739897 | 0.19 |
ENST00000605618.1 ENST00000427250.1 ENST00000601623.1 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr19_+_35739782 | 0.18 |
ENST00000347609.4 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr19_-_55652290 | 0.17 |
ENST00000589745.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr1_-_55266926 | 0.09 |
ENST00000371276.4 |
TTC22 |
tetratricopeptide repeat domain 22 |
| chr17_+_48585745 | 0.07 |
ENST00000323776.5 |
MYCBPAP |
MYCBP associated protein |
| chr6_+_53659746 | 0.07 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr11_-_58345569 | 0.06 |
ENST00000528954.1 ENST00000528489.1 |
LPXN |
leupaxin |
| chr6_+_18155560 | 0.05 |
ENST00000546309.2 ENST00000388870.2 ENST00000397244.1 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chrX_+_128913906 | 0.05 |
ENST00000356892.3 |
SASH3 |
SAM and SH3 domain containing 3 |
| chr7_+_1748798 | 0.03 |
ENST00000561626.1 |
ELFN1 |
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr6_+_116601265 | 0.03 |
ENST00000452085.3 |
DSE |
dermatan sulfate epimerase |
| chr6_-_26056695 | 0.03 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chr17_+_34136459 | 0.03 |
ENST00000588240.1 ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr1_-_55266865 | 0.03 |
ENST00000371274.4 |
TTC22 |
tetratricopeptide repeat domain 22 |
| chr6_+_29691198 | 0.03 |
ENST00000440587.2 ENST00000434407.2 |
HLA-F |
major histocompatibility complex, class I, F |
| chr2_-_172290482 | 0.03 |
ENST00000442541.1 ENST00000392599.2 ENST00000375258.4 |
METTL8 |
methyltransferase like 8 |
| chr11_+_13690249 | 0.03 |
ENST00000532701.1 |
FAR1 |
fatty acyl CoA reductase 1 |
| chr22_+_37415700 | 0.03 |
ENST00000397129.1 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr22_+_37415676 | 0.03 |
ENST00000401419.3 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr22_+_37415728 | 0.03 |
ENST00000404802.3 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr6_+_29691056 | 0.02 |
ENST00000414333.1 ENST00000334668.4 ENST00000259951.7 |
HLA-F |
major histocompatibility complex, class I, F |
| chr2_-_105946491 | 0.02 |
ENST00000393359.2 |
TGFBRAP1 |
transforming growth factor, beta receptor associated protein 1 |
| chr22_+_37415776 | 0.02 |
ENST00000341116.3 ENST00000429360.2 ENST00000404393.1 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr4_+_24797085 | 0.02 |
ENST00000382120.3 |
SOD3 |
superoxide dismutase 3, extracellular |
| chr1_-_246670519 | 0.02 |
ENST00000388985.4 ENST00000490107.1 |
SMYD3 |
SET and MYND domain containing 3 |
| chr2_+_172290707 | 0.02 |
ENST00000375255.3 ENST00000539783.1 |
DCAF17 |
DDB1 and CUL4 associated factor 17 |
| chrX_+_129535937 | 0.02 |
ENST00000305536.6 ENST00000370947.1 |
RBMX2 |
RNA binding motif protein, X-linked 2 |
| chr10_+_1034646 | 0.02 |
ENST00000360059.5 ENST00000545048.1 |
GTPBP4 |
GTP binding protein 4 |
| chr1_-_100715372 | 0.01 |
ENST00000370131.3 ENST00000370132.4 |
DBT |
dihydrolipoamide branched chain transacylase E2 |
| chr11_+_13690200 | 0.01 |
ENST00000354817.3 |
FAR1 |
fatty acyl CoA reductase 1 |
| chr7_-_74489609 | 0.01 |
ENST00000329959.4 ENST00000503250.2 ENST00000543840.1 |
WBSCR16 |
Williams-Beuren syndrome chromosome region 16 |
| chr16_+_21964662 | 0.01 |
ENST00000561553.1 ENST00000565331.1 |
UQCRC2 |
ubiquinol-cytochrome c reductase core protein II |
| chr8_+_145064233 | 0.01 |
ENST00000529301.1 ENST00000395068.4 |
GRINA |
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr6_-_31763408 | 0.01 |
ENST00000444930.2 |
VARS |
valyl-tRNA synthetase |
| chr8_-_109260897 | 0.01 |
ENST00000521297.1 ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E |
eukaryotic translation initiation factor 3, subunit E |
| chr7_+_117824086 | 0.01 |
ENST00000249299.2 ENST00000424702.1 |
NAA38 |
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr6_+_31939608 | 0.01 |
ENST00000375331.2 ENST00000375333.2 |
STK19 |
serine/threonine kinase 19 |
| chr17_-_27169745 | 0.01 |
ENST00000583307.1 ENST00000581229.1 ENST00000582266.1 ENST00000577376.1 ENST00000577682.1 ENST00000581381.1 ENST00000341217.5 ENST00000581407.1 ENST00000583522.1 |
FAM222B |
family with sequence similarity 222, member B |
| chr1_-_235292250 | 0.01 |
ENST00000366607.4 |
TOMM20 |
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr5_+_138677515 | 0.01 |
ENST00000265192.4 ENST00000511706.1 |
PAIP2 |
poly(A) binding protein interacting protein 2 |
| chr1_+_214161272 | 0.01 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr17_-_45908875 | 0.01 |
ENST00000351111.2 ENST00000414011.1 |
MRPL10 |
mitochondrial ribosomal protein L10 |
| chr7_+_100303676 | 0.01 |
ENST00000303151.4 |
POP7 |
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr14_+_51706886 | 0.01 |
ENST00000457354.2 |
TMX1 |
thioredoxin-related transmembrane protein 1 |
| chr12_-_46662772 | 0.01 |
ENST00000549049.1 ENST00000439706.1 ENST00000398637.5 |
SLC38A1 |
solute carrier family 38, member 1 |
| chrX_+_1387693 | 0.01 |
ENST00000381529.3 ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr11_+_46638805 | 0.01 |
ENST00000434074.1 ENST00000312040.4 ENST00000451945.1 |
ATG13 |
autophagy related 13 |
| chr15_+_85523671 | 0.01 |
ENST00000310298.4 ENST00000557957.1 |
PDE8A |
phosphodiesterase 8A |
| chr12_-_74686314 | 0.00 |
ENST00000551210.1 ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2 |
RP11-81H3.2 |
| chr22_+_30163340 | 0.00 |
ENST00000330029.6 ENST00000401406.3 |
UQCR10 |
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr9_+_96026230 | 0.00 |
ENST00000448251.1 |
WNK2 |
WNK lysine deficient protein kinase 2 |
| chr2_-_240322643 | 0.00 |
ENST00000345617.3 |
HDAC4 |
histone deacetylase 4 |
| chr11_+_46402583 | 0.00 |
ENST00000359803.3 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr13_-_21750659 | 0.00 |
ENST00000400018.3 ENST00000314759.5 |
SKA3 |
spindle and kinetochore associated complex subunit 3 |
| chr2_+_198365122 | 0.00 |
ENST00000604458.1 |
HSPE1-MOB4 |
HSPE1-MOB4 readthrough |
| chr22_-_30162924 | 0.00 |
ENST00000344318.3 ENST00000397781.3 |
ZMAT5 |
zinc finger, matrin-type 5 |
| chr12_+_52463751 | 0.00 |
ENST00000336854.4 ENST00000550604.1 ENST00000553049.1 ENST00000548915.1 |
C12orf44 |
chromosome 12 open reading frame 44 |
| chr6_+_88117683 | 0.00 |
ENST00000369562.4 |
C6ORF165 |
UPF0704 protein C6orf165 |
| chr19_-_5903714 | 0.00 |
ENST00000586349.1 ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12 NDUFA11 FUT5 |
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr13_+_21750780 | 0.00 |
ENST00000309594.4 |
MRP63 |
mitochondrial ribosomal protein 63 |
| chr19_+_37998031 | 0.00 |
ENST00000586138.1 ENST00000588578.1 ENST00000587986.1 |
ZNF793 |
zinc finger protein 793 |
| chr8_+_61429416 | 0.00 |
ENST00000262646.7 ENST00000531289.1 |
RAB2A |
RAB2A, member RAS oncogene family |
| chr12_-_122750957 | 0.00 |
ENST00000451053.2 |
VPS33A |
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr16_+_68573116 | 0.00 |
ENST00000570495.1 ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90 |
ZFP90 zinc finger protein |
| chr9_-_32552551 | 0.00 |
ENST00000360538.2 ENST00000379858.1 |
TOPORS |
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.1 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |