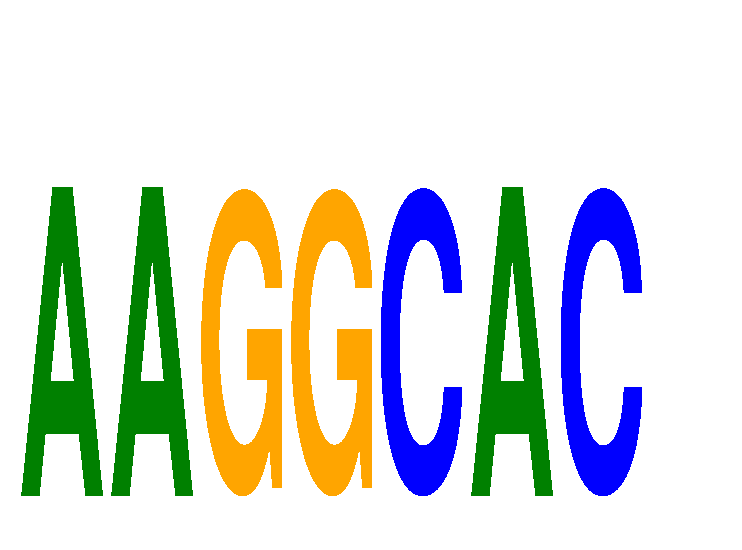Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for AAGGCAC
Z-value: 0.79
miRNA associated with seed AAGGCAC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-124-3p.1
|
MIMAT0000422 |
Activity profile of AAGGCAC motif
Sorted Z-values of AAGGCAC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGCAC
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_121513143 | 0.97 |
ENST00000393386.2 |
PTPRZ1 |
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr14_-_105635090 | 0.93 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr12_+_41086297 | 0.85 |
ENST00000551295.2 |
CNTN1 |
contactin 1 |
| chr1_-_242687676 | 0.84 |
ENST00000536534.2 |
PLD5 |
phospholipase D family, member 5 |
| chr3_-_69435224 | 0.84 |
ENST00000398540.3 |
FRMD4B |
FERM domain containing 4B |
| chr6_-_136871957 | 0.74 |
ENST00000354570.3 |
MAP7 |
microtubule-associated protein 7 |
| chr1_+_151483855 | 0.72 |
ENST00000427934.2 ENST00000271636.7 |
CGN |
cingulin |
| chr1_+_92495528 | 0.71 |
ENST00000370383.4 |
EPHX4 |
epoxide hydrolase 4 |
| chr9_+_115913222 | 0.70 |
ENST00000259392.3 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
| chr18_+_19749386 | 0.70 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr8_-_15095832 | 0.69 |
ENST00000382080.1 |
SGCZ |
sarcoglycan, zeta |
| chr10_+_95653687 | 0.69 |
ENST00000371408.3 ENST00000427197.1 |
SLC35G1 |
solute carrier family 35, member G1 |
| chr1_-_21059029 | 0.69 |
ENST00000444387.2 ENST00000375031.1 ENST00000518294.1 |
SH2D5 |
SH2 domain containing 5 |
| chr14_-_99737565 | 0.66 |
ENST00000357195.3 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr1_-_201368707 | 0.63 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr20_-_14318248 | 0.62 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr11_-_115375107 | 0.61 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chrX_+_105937068 | 0.61 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr19_+_54371114 | 0.60 |
ENST00000448420.1 ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM |
myeloid-associated differentiation marker |
| chr12_+_4382917 | 0.58 |
ENST00000261254.3 |
CCND2 |
cyclin D2 |
| chr5_-_16509101 | 0.58 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr18_-_47721447 | 0.58 |
ENST00000285039.7 |
MYO5B |
myosin VB |
| chr19_-_10697895 | 0.57 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr10_-_105615164 | 0.56 |
ENST00000355946.2 ENST00000369774.4 |
SH3PXD2A |
SH3 and PX domains 2A |
| chr6_+_1312675 | 0.56 |
ENST00000296839.2 |
FOXQ1 |
forkhead box Q1 |
| chr20_-_18038521 | 0.55 |
ENST00000278780.6 |
OVOL2 |
ovo-like zinc finger 2 |
| chr2_-_122042770 | 0.55 |
ENST00000263707.5 |
TFCP2L1 |
transcription factor CP2-like 1 |
| chr14_+_67999999 | 0.53 |
ENST00000329153.5 |
PLEKHH1 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr11_+_10471836 | 0.53 |
ENST00000444303.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr4_-_80994210 | 0.52 |
ENST00000403729.2 |
ANTXR2 |
anthrax toxin receptor 2 |
| chr12_-_95044309 | 0.51 |
ENST00000261226.4 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
| chr13_-_86373536 | 0.50 |
ENST00000400286.2 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
| chr1_+_26856236 | 0.49 |
ENST00000374168.2 ENST00000374166.4 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr9_+_22446814 | 0.47 |
ENST00000325870.2 |
DMRTA1 |
DMRT-like family A1 |
| chr2_+_26568965 | 0.47 |
ENST00000260585.7 ENST00000447170.1 |
EPT1 |
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr15_-_79237433 | 0.47 |
ENST00000220166.5 |
CTSH |
cathepsin H |
| chr2_-_208634287 | 0.44 |
ENST00000295417.3 |
FZD5 |
frizzled family receptor 5 |
| chr4_+_95679072 | 0.44 |
ENST00000515059.1 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chrX_+_68048803 | 0.43 |
ENST00000204961.4 |
EFNB1 |
ephrin-B1 |
| chr8_+_61591337 | 0.42 |
ENST00000423902.2 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
| chr2_+_112812778 | 0.42 |
ENST00000283206.4 |
TMEM87B |
transmembrane protein 87B |
| chr18_+_29598335 | 0.42 |
ENST00000217740.3 |
RNF125 |
ring finger protein 125, E3 ubiquitin protein ligase |
| chr17_-_47841485 | 0.39 |
ENST00000506156.1 ENST00000240364.2 |
FAM117A |
family with sequence similarity 117, member A |
| chr8_-_145013711 | 0.38 |
ENST00000345136.3 |
PLEC |
plectin |
| chr1_+_31885963 | 0.38 |
ENST00000373709.3 |
SERINC2 |
serine incorporator 2 |
| chr6_+_53659746 | 0.37 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr8_-_75233563 | 0.37 |
ENST00000342232.4 |
JPH1 |
junctophilin 1 |
| chr9_-_135996537 | 0.37 |
ENST00000372050.3 ENST00000372047.3 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
| chr1_-_116311402 | 0.37 |
ENST00000261448.5 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
| chr15_-_59665062 | 0.36 |
ENST00000288235.4 |
MYO1E |
myosin IE |
| chr12_+_65563329 | 0.36 |
ENST00000308330.2 |
LEMD3 |
LEM domain containing 3 |
| chr6_+_37137939 | 0.36 |
ENST00000373509.5 |
PIM1 |
pim-1 oncogene |
| chr10_-_35930219 | 0.36 |
ENST00000374694.1 |
FZD8 |
frizzled family receptor 8 |
| chr3_+_36421826 | 0.36 |
ENST00000273183.3 |
STAC |
SH3 and cysteine rich domain |
| chr1_-_226924980 | 0.35 |
ENST00000272117.3 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr14_+_65171099 | 0.34 |
ENST00000247226.7 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr8_+_74206829 | 0.34 |
ENST00000240285.5 |
RDH10 |
retinol dehydrogenase 10 (all-trans) |
| chr10_+_71211212 | 0.34 |
ENST00000373290.2 |
TSPAN15 |
tetraspanin 15 |
| chr22_+_29279552 | 0.34 |
ENST00000544604.2 |
ZNRF3 |
zinc and ring finger 3 |
| chr2_-_9143786 | 0.34 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr3_-_185216766 | 0.33 |
ENST00000296254.3 |
TMEM41A |
transmembrane protein 41A |
| chr9_+_470288 | 0.33 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr14_-_23652849 | 0.33 |
ENST00000316902.7 ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr2_+_70142189 | 0.33 |
ENST00000264444.2 |
MXD1 |
MAX dimerization protein 1 |
| chr6_+_7727030 | 0.33 |
ENST00000283147.6 |
BMP6 |
bone morphogenetic protein 6 |
| chr6_+_44187242 | 0.33 |
ENST00000393844.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr11_-_87908600 | 0.33 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chr14_+_70078303 | 0.32 |
ENST00000342745.4 |
KIAA0247 |
KIAA0247 |
| chr1_-_19283163 | 0.32 |
ENST00000455833.2 |
IFFO2 |
intermediate filament family orphan 2 |
| chr9_+_2621798 | 0.32 |
ENST00000382100.3 |
VLDLR |
very low density lipoprotein receptor |
| chr16_-_402639 | 0.32 |
ENST00000262320.3 |
AXIN1 |
axin 1 |
| chr19_+_38924316 | 0.31 |
ENST00000355481.4 ENST00000360985.3 ENST00000359596.3 |
RYR1 |
ryanodine receptor 1 (skeletal) |
| chr1_+_160370344 | 0.31 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr6_+_12012536 | 0.31 |
ENST00000379388.2 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
| chr14_-_95786200 | 0.31 |
ENST00000298912.4 |
CLMN |
calmin (calponin-like, transmembrane) |
| chr7_-_105162652 | 0.31 |
ENST00000356362.2 ENST00000469408.1 |
PUS7 |
pseudouridylate synthase 7 homolog (S. cerevisiae) |
| chr8_-_66754172 | 0.30 |
ENST00000401827.3 |
PDE7A |
phosphodiesterase 7A |
| chr1_+_955448 | 0.30 |
ENST00000379370.2 |
AGRN |
agrin |
| chr12_+_5019061 | 0.30 |
ENST00000382545.3 |
KCNA1 |
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) |
| chr1_+_65613217 | 0.30 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr5_+_140220769 | 0.30 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chrX_-_33146477 | 0.30 |
ENST00000378677.2 |
DMD |
dystrophin |
| chr8_+_6565854 | 0.30 |
ENST00000285518.6 |
AGPAT5 |
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr5_+_140213815 | 0.30 |
ENST00000525929.1 ENST00000378125.3 |
PCDHA7 |
protocadherin alpha 7 |
| chr5_+_140227048 | 0.30 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr5_+_140254884 | 0.30 |
ENST00000398631.2 |
PCDHA12 |
protocadherin alpha 12 |
| chr2_+_206547215 | 0.30 |
ENST00000360409.3 ENST00000540178.1 ENST00000540841.1 ENST00000355117.4 ENST00000450507.1 ENST00000417189.1 |
NRP2 |
neuropilin 2 |
| chr8_-_494824 | 0.30 |
ENST00000427263.2 ENST00000324079.6 |
TDRP |
testis development related protein |
| chr5_+_140248518 | 0.29 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chr18_+_29077990 | 0.29 |
ENST00000261590.8 |
DSG2 |
desmoglein 2 |
| chr1_+_220701456 | 0.29 |
ENST00000366918.4 ENST00000402574.1 |
MARK1 |
MAP/microtubule affinity-regulating kinase 1 |
| chr10_-_52383644 | 0.29 |
ENST00000361781.2 |
SGMS1 |
sphingomyelin synthase 1 |
| chr3_+_5229356 | 0.29 |
ENST00000256497.4 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
| chr1_-_113498943 | 0.29 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr13_+_31774073 | 0.28 |
ENST00000343307.4 |
B3GALTL |
beta 1,3-galactosyltransferase-like |
| chr9_-_124132483 | 0.28 |
ENST00000286713.2 ENST00000538954.1 ENST00000347359.2 |
STOM |
stomatin |
| chr7_+_89841000 | 0.28 |
ENST00000287908.3 |
STEAP2 |
STEAP family member 2, metalloreductase |
| chr15_+_73976545 | 0.28 |
ENST00000318443.5 ENST00000537340.2 ENST00000318424.5 |
CD276 |
CD276 molecule |
| chr11_+_76494253 | 0.28 |
ENST00000333090.4 |
TSKU |
tsukushi, small leucine rich proteoglycan |
| chr11_+_120081475 | 0.27 |
ENST00000328965.4 |
OAF |
OAF homolog (Drosophila) |
| chr15_+_33603147 | 0.27 |
ENST00000415757.3 ENST00000389232.4 |
RYR3 |
ryanodine receptor 3 |
| chr14_-_78083112 | 0.27 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr11_+_65554493 | 0.26 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr12_+_44229846 | 0.26 |
ENST00000551577.1 ENST00000266534.3 |
TMEM117 |
transmembrane protein 117 |
| chr9_+_101569944 | 0.26 |
ENST00000375011.3 |
GALNT12 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) |
| chr7_-_108096822 | 0.26 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr11_+_118230287 | 0.26 |
ENST00000252108.3 ENST00000431736.2 |
UBE4A |
ubiquitination factor E4A |
| chr17_-_17494972 | 0.26 |
ENST00000435340.2 ENST00000255389.5 ENST00000395781.2 |
PEMT |
phosphatidylethanolamine N-methyltransferase |
| chr13_+_80055284 | 0.26 |
ENST00000218652.7 |
NDFIP2 |
Nedd4 family interacting protein 2 |
| chr11_+_121322832 | 0.26 |
ENST00000260197.7 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr6_+_7107999 | 0.25 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr16_-_17564738 | 0.25 |
ENST00000261381.6 |
XYLT1 |
xylosyltransferase I |
| chr13_-_30169807 | 0.25 |
ENST00000380752.5 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr8_-_57906362 | 0.25 |
ENST00000262644.4 |
IMPAD1 |
inositol monophosphatase domain containing 1 |
| chr6_-_33756867 | 0.25 |
ENST00000293760.5 |
LEMD2 |
LEM domain containing 2 |
| chr19_+_39138271 | 0.25 |
ENST00000252699.2 |
ACTN4 |
actinin, alpha 4 |
| chr7_-_100860851 | 0.25 |
ENST00000223127.3 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr1_+_205473720 | 0.25 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr22_+_50624323 | 0.25 |
ENST00000380909.4 ENST00000303434.4 |
TRABD |
TraB domain containing |
| chr3_-_27498235 | 0.24 |
ENST00000295736.5 ENST00000428386.1 ENST00000428179.1 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr2_-_97405775 | 0.24 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr20_+_361261 | 0.24 |
ENST00000217233.3 |
TRIB3 |
tribbles pseudokinase 3 |
| chr18_+_42260861 | 0.24 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr7_-_112430647 | 0.24 |
ENST00000312814.6 |
TMEM168 |
transmembrane protein 168 |
| chr17_-_8151353 | 0.24 |
ENST00000315684.8 |
CTC1 |
CTS telomere maintenance complex component 1 |
| chr7_+_24612935 | 0.24 |
ENST00000222644.5 |
MPP6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr10_+_98741041 | 0.24 |
ENST00000286067.2 |
C10orf12 |
chromosome 10 open reading frame 12 |
| chr9_-_34637718 | 0.24 |
ENST00000378892.1 ENST00000277010.4 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr13_-_21476900 | 0.24 |
ENST00000400602.2 ENST00000255305.6 |
XPO4 |
exportin 4 |
| chr6_-_107436473 | 0.24 |
ENST00000369042.1 |
BEND3 |
BEN domain containing 3 |
| chr18_-_19284724 | 0.24 |
ENST00000580981.1 ENST00000289119.2 |
ABHD3 |
abhydrolase domain containing 3 |
| chr7_-_124405681 | 0.24 |
ENST00000303921.2 |
GPR37 |
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr9_-_34376851 | 0.23 |
ENST00000297625.7 |
KIAA1161 |
KIAA1161 |
| chr2_+_118846008 | 0.23 |
ENST00000245787.4 |
INSIG2 |
insulin induced gene 2 |
| chr12_-_57472522 | 0.23 |
ENST00000379391.3 ENST00000300128.4 |
TMEM194A |
transmembrane protein 194A |
| chr11_+_60681346 | 0.23 |
ENST00000227525.3 |
TMEM109 |
transmembrane protein 109 |
| chr16_-_68482440 | 0.23 |
ENST00000219334.5 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr19_-_45908292 | 0.23 |
ENST00000360957.5 ENST00000592134.1 |
PPP1R13L |
protein phosphatase 1, regulatory subunit 13 like |
| chr11_+_75479763 | 0.22 |
ENST00000228027.7 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
| chr18_-_53255766 | 0.22 |
ENST00000566286.1 ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4 |
transcription factor 4 |
| chr11_+_7597639 | 0.22 |
ENST00000533792.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr14_-_64194745 | 0.22 |
ENST00000247225.6 |
SGPP1 |
sphingosine-1-phosphate phosphatase 1 |
| chr10_-_131762105 | 0.22 |
ENST00000368648.3 ENST00000355311.5 |
EBF3 |
early B-cell factor 3 |
| chr22_-_36784035 | 0.22 |
ENST00000216181.5 |
MYH9 |
myosin, heavy chain 9, non-muscle |
| chr8_+_32405728 | 0.22 |
ENST00000523079.1 ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1 |
neuregulin 1 |
| chr5_-_16936340 | 0.21 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr12_+_133066137 | 0.21 |
ENST00000434748.2 |
FBRSL1 |
fibrosin-like 1 |
| chr19_+_797392 | 0.21 |
ENST00000350092.4 ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1 |
polypyrimidine tract binding protein 1 |
| chr20_-_50419055 | 0.21 |
ENST00000217086.4 |
SALL4 |
spalt-like transcription factor 4 |
| chr19_-_55954230 | 0.21 |
ENST00000376325.4 |
SHISA7 |
shisa family member 7 |
| chr8_-_20161466 | 0.21 |
ENST00000381569.1 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
| chr12_-_129308487 | 0.21 |
ENST00000266771.5 |
SLC15A4 |
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr4_+_108745711 | 0.21 |
ENST00000394684.4 |
SGMS2 |
sphingomyelin synthase 2 |
| chr5_+_56111361 | 0.21 |
ENST00000399503.3 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr6_+_73331776 | 0.21 |
ENST00000370398.1 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr5_+_149109825 | 0.21 |
ENST00000360453.4 ENST00000394320.3 ENST00000309241.5 |
PPARGC1B |
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr10_-_106098162 | 0.20 |
ENST00000337478.1 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr9_+_116638562 | 0.20 |
ENST00000374126.5 ENST00000288466.7 |
ZNF618 |
zinc finger protein 618 |
| chr17_-_74449252 | 0.20 |
ENST00000319380.7 |
UBE2O |
ubiquitin-conjugating enzyme E2O |
| chr22_+_21996549 | 0.20 |
ENST00000248958.4 |
SDF2L1 |
stromal cell-derived factor 2-like 1 |
| chr1_-_169455169 | 0.20 |
ENST00000367804.4 ENST00000236137.5 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
| chr10_+_72575643 | 0.20 |
ENST00000373202.3 |
SGPL1 |
sphingosine-1-phosphate lyase 1 |
| chr16_-_2246436 | 0.20 |
ENST00000343516.6 |
CASKIN1 |
CASK interacting protein 1 |
| chr2_+_103236004 | 0.20 |
ENST00000233969.2 |
SLC9A2 |
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr2_+_191045562 | 0.20 |
ENST00000340623.4 |
C2orf88 |
chromosome 2 open reading frame 88 |
| chr5_+_109025067 | 0.20 |
ENST00000261483.4 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
| chr11_+_20620946 | 0.20 |
ENST00000525748.1 |
SLC6A5 |
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr14_+_71108460 | 0.20 |
ENST00000256367.2 |
TTC9 |
tetratricopeptide repeat domain 9 |
| chr11_-_118023490 | 0.20 |
ENST00000324727.4 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
| chr1_+_116184566 | 0.20 |
ENST00000355485.2 ENST00000369510.4 |
VANGL1 |
VANGL planar cell polarity protein 1 |
| chr5_+_140345820 | 0.19 |
ENST00000289269.5 |
PCDHAC2 |
protocadherin alpha subfamily C, 2 |
| chr1_-_204329013 | 0.19 |
ENST00000272203.3 ENST00000414478.1 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
| chr5_+_127419449 | 0.19 |
ENST00000262461.2 ENST00000343225.4 |
SLC12A2 |
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr11_+_13690200 | 0.19 |
ENST00000354817.3 |
FAR1 |
fatty acyl CoA reductase 1 |
| chr8_+_98881268 | 0.19 |
ENST00000254898.5 ENST00000524308.1 ENST00000522025.2 |
MATN2 |
matrilin 2 |
| chr19_-_1174226 | 0.19 |
ENST00000587024.1 ENST00000361757.3 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
| chr11_+_60609537 | 0.19 |
ENST00000227520.5 |
CCDC86 |
coiled-coil domain containing 86 |
| chr12_-_93323013 | 0.19 |
ENST00000322349.8 |
EEA1 |
early endosome antigen 1 |
| chr11_-_59383617 | 0.19 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr3_-_4508925 | 0.19 |
ENST00000534863.1 ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1 |
sulfatase modifying factor 1 |
| chr1_+_179050512 | 0.19 |
ENST00000367627.3 |
TOR3A |
torsin family 3, member A |
| chr6_-_43543702 | 0.19 |
ENST00000265351.7 |
XPO5 |
exportin 5 |
| chr19_-_39805976 | 0.19 |
ENST00000248668.4 |
LRFN1 |
leucine rich repeat and fibronectin type III domain containing 1 |
| chr11_+_68816356 | 0.19 |
ENST00000294309.3 ENST00000542467.1 |
TPCN2 |
two pore segment channel 2 |
| chr2_+_134877740 | 0.19 |
ENST00000409645.1 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr8_+_98656336 | 0.19 |
ENST00000336273.3 |
MTDH |
metadherin |
| chr1_+_40723779 | 0.19 |
ENST00000372759.3 |
ZMPSTE24 |
zinc metallopeptidase STE24 |
| chr9_+_97136833 | 0.19 |
ENST00000375344.3 |
HIATL1 |
hippocampus abundant transcript-like 1 |
| chr11_+_64126614 | 0.19 |
ENST00000528057.1 ENST00000334205.4 ENST00000294261.4 |
RPS6KA4 |
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr4_+_152330390 | 0.18 |
ENST00000503146.1 ENST00000435205.1 |
FAM160A1 |
family with sequence similarity 160, member A1 |
| chr22_+_50247449 | 0.18 |
ENST00000216268.5 |
ZBED4 |
zinc finger, BED-type containing 4 |
| chr20_-_43977055 | 0.18 |
ENST00000372733.3 ENST00000537976.1 |
SDC4 |
syndecan 4 |
| chr7_+_77166592 | 0.18 |
ENST00000248594.6 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
| chr6_-_53213780 | 0.18 |
ENST00000304434.6 ENST00000370918.4 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
| chr16_+_55542910 | 0.18 |
ENST00000262134.5 |
LPCAT2 |
lysophosphatidylcholine acyltransferase 2 |
| chr5_+_140207536 | 0.18 |
ENST00000529310.1 ENST00000527624.1 |
PCDHA6 |
protocadherin alpha 6 |
| chr5_-_115910630 | 0.18 |
ENST00000343348.6 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_-_160472952 | 0.17 |
ENST00000541068.2 ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr4_-_100485143 | 0.17 |
ENST00000394877.3 |
TRMT10A |
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr12_-_7125770 | 0.17 |
ENST00000261407.4 |
LPCAT3 |
lysophosphatidylcholine acyltransferase 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 0.9 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.2 | 0.5 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 0.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:1901382 | Spemann organizer formation(GO:0060061) chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.7 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.9 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.9 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.1 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.1 | 0.3 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 0.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.5 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.3 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.4 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.5 | GO:0003160 | endocardium morphogenesis(GO:0003160) endocardium formation(GO:0060214) |
| 0.1 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.6 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.3 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.1 | 0.3 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.1 | 0.4 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.8 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.3 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.3 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.1 | 0.9 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.6 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.7 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.3 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.1 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.2 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.6 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.5 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 0.2 | GO:1900082 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.2 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.4 | GO:0097461 | copper ion import(GO:0015677) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.2 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.2 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.2 | GO:1902683 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.3 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0055048 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.5 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.4 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.3 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.2 | GO:0090175 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0048377 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) mesendoderm development(GO:0048382) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.2 | GO:0031944 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of hormone biosynthetic process(GO:0032353) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.0 | 0.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0090210 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.6 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.4 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0097491 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.3 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 | 0.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.7 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 3.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.5 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.0 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0051643 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) secretory granule localization(GO:0032252) endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.1 | GO:0048617 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 0.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 1.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.3 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.2 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 0.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.0 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 1.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.3 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.3 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.3 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.3 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.2 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.2 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 1.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.0 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.0 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.5 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |