Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for AIRE
Z-value: 0.68
Transcription factors associated with AIRE
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AIRE
|
ENSG00000160224.12 | AIRE |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AIRE | hg19_v2_chr21_+_45705752_45705767 | 0.16 | 7.0e-01 | Click! |
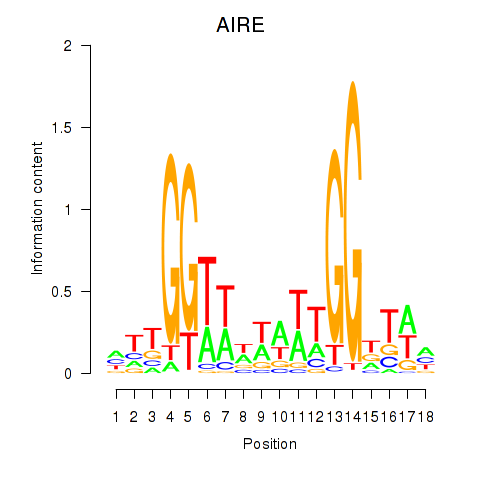
Activity profile of AIRE motif
Sorted Z-values of AIRE motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AIRE
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_90112117 | 0.96 |
ENST00000358077.5 |
DAPK1 |
death-associated protein kinase 1 |
| chr12_-_15038779 | 0.94 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr2_-_225811747 | 0.85 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr4_-_57524061 | 0.81 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chr8_+_104384616 | 0.77 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr12_+_75874984 | 0.67 |
ENST00000550491.1 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr20_+_12989596 | 0.55 |
ENST00000434210.1 ENST00000399002.2 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
| chr6_-_31514333 | 0.53 |
ENST00000376151.4 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr5_+_32788945 | 0.52 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr12_+_75874580 | 0.49 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr4_-_186696425 | 0.48 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr1_-_243326612 | 0.46 |
ENST00000492145.1 ENST00000490813.1 ENST00000464936.1 |
CEP170 |
centrosomal protein 170kDa |
| chr6_-_31514516 | 0.45 |
ENST00000303892.5 ENST00000483251.1 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr12_+_75874460 | 0.42 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr12_+_55248289 | 0.42 |
ENST00000308796.6 |
MUCL1 |
mucin-like 1 |
| chr9_-_113761720 | 0.37 |
ENST00000541779.1 ENST00000374430.2 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr9_-_21482312 | 0.37 |
ENST00000448696.3 |
IFNE |
interferon, epsilon |
| chr10_+_90660832 | 0.35 |
ENST00000371924.1 |
STAMBPL1 |
STAM binding protein-like 1 |
| chr19_-_52551814 | 0.31 |
ENST00000594154.1 ENST00000598745.1 ENST00000597273.1 |
ZNF432 |
zinc finger protein 432 |
| chr1_+_79115503 | 0.29 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr5_+_72143988 | 0.26 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chrX_-_100307076 | 0.26 |
ENST00000338687.7 ENST00000545398.1 ENST00000372931.5 |
TRMT2B |
tRNA methyltransferase 2 homolog B (S. cerevisiae) |
| chr20_-_33735070 | 0.25 |
ENST00000374491.3 ENST00000542871.1 ENST00000374492.3 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chrX_-_100307043 | 0.25 |
ENST00000372939.1 ENST00000372935.1 ENST00000372936.3 |
TRMT2B |
tRNA methyltransferase 2 homolog B (S. cerevisiae) |
| chrM_+_12331 | 0.24 |
ENST00000361567.2 |
MT-ND5 |
mitochondrially encoded NADH dehydrogenase 5 |
| chr3_-_49466686 | 0.24 |
ENST00000273598.3 ENST00000436744.2 |
NICN1 |
nicolin 1 |
| chr10_-_106240032 | 0.24 |
ENST00000447860.1 |
RP11-127O4.3 |
RP11-127O4.3 |
| chr5_+_140625147 | 0.23 |
ENST00000231173.3 |
PCDHB15 |
protocadherin beta 15 |
| chr19_+_11455900 | 0.23 |
ENST00000588790.1 |
CCDC159 |
coiled-coil domain containing 159 |
| chr19_-_48752812 | 0.22 |
ENST00000359009.4 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr6_+_36839616 | 0.22 |
ENST00000359359.2 ENST00000510325.2 |
C6orf89 |
chromosome 6 open reading frame 89 |
| chr2_-_166060571 | 0.22 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr12_-_6580094 | 0.22 |
ENST00000361716.3 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr13_+_50570019 | 0.22 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr19_-_48753104 | 0.21 |
ENST00000447740.2 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr1_+_174843548 | 0.21 |
ENST00000478442.1 ENST00000465412.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr4_-_114682936 | 0.20 |
ENST00000454265.2 ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr2_-_166060552 | 0.20 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr16_+_32077386 | 0.18 |
ENST00000354689.6 |
IGHV3OR16-9 |
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr9_-_114361665 | 0.17 |
ENST00000309195.5 |
PTGR1 |
prostaglandin reductase 1 |
| chr8_-_7220490 | 0.17 |
ENST00000400078.2 |
ZNF705G |
zinc finger protein 705G |
| chr4_-_66536057 | 0.17 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr15_+_27112948 | 0.17 |
ENST00000555060.1 |
GABRA5 |
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr17_-_295730 | 0.16 |
ENST00000329099.4 |
FAM101B |
family with sequence similarity 101, member B |
| chr19_+_18208603 | 0.16 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr19_-_23869999 | 0.16 |
ENST00000601935.1 ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675 |
zinc finger protein 675 |
| chr12_-_6579808 | 0.15 |
ENST00000535180.1 ENST00000400911.3 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr9_-_114362093 | 0.15 |
ENST00000538962.1 ENST00000407693.2 ENST00000238248.3 |
PTGR1 |
prostaglandin reductase 1 |
| chr2_+_179316163 | 0.14 |
ENST00000409117.3 |
DFNB59 |
deafness, autosomal recessive 59 |
| chr2_+_201981527 | 0.14 |
ENST00000441224.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr7_-_87936195 | 0.14 |
ENST00000414498.1 ENST00000301959.5 ENST00000380079.4 |
STEAP4 |
STEAP family member 4 |
| chr10_-_101841588 | 0.14 |
ENST00000370418.3 |
CPN1 |
carboxypeptidase N, polypeptide 1 |
| chr9_-_114361919 | 0.13 |
ENST00000422125.1 |
PTGR1 |
prostaglandin reductase 1 |
| chr5_-_107703556 | 0.13 |
ENST00000496714.1 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
| chr9_-_4666421 | 0.13 |
ENST00000381895.5 |
SPATA6L |
spermatogenesis associated 6-like |
| chr14_-_24911971 | 0.11 |
ENST00000555365.1 ENST00000399395.3 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr8_+_105352050 | 0.11 |
ENST00000297581.2 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
| chr12_+_52643077 | 0.11 |
ENST00000553310.2 ENST00000544024.1 |
KRT86 |
keratin 86 |
| chr16_+_14802801 | 0.11 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr16_+_3254247 | 0.11 |
ENST00000304646.2 |
OR1F1 |
olfactory receptor, family 1, subfamily F, member 1 |
| chr14_+_58894706 | 0.11 |
ENST00000261244.5 |
KIAA0586 |
KIAA0586 |
| chr1_-_145826450 | 0.11 |
ENST00000462900.2 |
GPR89A |
G protein-coupled receptor 89A |
| chr3_-_19975665 | 0.10 |
ENST00000295824.9 ENST00000389256.4 |
EFHB |
EF-hand domain family, member B |
| chr1_+_152627927 | 0.10 |
ENST00000444515.1 ENST00000536536.1 |
LINC00302 |
long intergenic non-protein coding RNA 302 |
| chr2_+_172864490 | 0.10 |
ENST00000315796.4 |
METAP1D |
methionyl aminopeptidase type 1D (mitochondrial) |
| chrM_-_14670 | 0.10 |
ENST00000361681.2 |
MT-ND6 |
mitochondrially encoded NADH dehydrogenase 6 |
| chr4_-_104119528 | 0.10 |
ENST00000380026.3 ENST00000503705.1 ENST00000265148.3 |
CENPE |
centromere protein E, 312kDa |
| chr2_-_202508169 | 0.10 |
ENST00000409883.2 |
TMEM237 |
transmembrane protein 237 |
| chr5_+_122181184 | 0.09 |
ENST00000513881.1 |
SNX24 |
sorting nexin 24 |
| chr16_-_71518504 | 0.09 |
ENST00000565100.2 |
ZNF19 |
zinc finger protein 19 |
| chr2_+_166150541 | 0.09 |
ENST00000283256.6 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr5_+_122181279 | 0.09 |
ENST00000395451.4 ENST00000506996.1 |
SNX24 |
sorting nexin 24 |
| chr22_+_40322595 | 0.09 |
ENST00000420971.1 ENST00000544756.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr15_+_52043758 | 0.09 |
ENST00000249700.4 ENST00000539962.2 |
TMOD2 |
tropomodulin 2 (neuronal) |
| chr19_-_23869970 | 0.09 |
ENST00000601010.1 |
ZNF675 |
zinc finger protein 675 |
| chr22_+_40322623 | 0.08 |
ENST00000399090.2 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr14_-_24912047 | 0.08 |
ENST00000553930.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr3_+_189507460 | 0.08 |
ENST00000434928.1 |
TP63 |
tumor protein p63 |
| chr11_+_30252549 | 0.08 |
ENST00000254122.3 ENST00000417547.1 |
FSHB |
follicle stimulating hormone, beta polypeptide |
| chr8_-_7309887 | 0.08 |
ENST00000458665.1 ENST00000528168.1 |
SPAG11B |
sperm associated antigen 11B |
| chr17_-_38721711 | 0.08 |
ENST00000578085.1 ENST00000246657.2 |
CCR7 |
chemokine (C-C motif) receptor 7 |
| chr14_-_55658323 | 0.07 |
ENST00000554067.1 ENST00000247191.2 |
DLGAP5 |
discs, large (Drosophila) homolog-associated protein 5 |
| chr13_+_103459704 | 0.07 |
ENST00000602836.1 |
BIVM-ERCC5 |
BIVM-ERCC5 readthrough |
| chr3_-_123710199 | 0.07 |
ENST00000184183.4 |
ROPN1 |
rhophilin associated tail protein 1 |
| chr20_-_43150601 | 0.07 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr5_+_122181128 | 0.07 |
ENST00000261369.4 |
SNX24 |
sorting nexin 24 |
| chr19_+_11708259 | 0.07 |
ENST00000587939.1 ENST00000588174.1 |
ZNF627 |
zinc finger protein 627 |
| chr11_+_64794875 | 0.07 |
ENST00000377244.3 ENST00000534637.1 ENST00000524831.1 |
SNX15 |
sorting nexin 15 |
| chr19_-_36643329 | 0.07 |
ENST00000589154.1 |
COX7A1 |
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr8_+_7716700 | 0.07 |
ENST00000454911.2 ENST00000326625.5 |
SPAG11A |
sperm associated antigen 11A |
| chr4_+_78829479 | 0.07 |
ENST00000504901.1 |
MRPL1 |
mitochondrial ribosomal protein L1 |
| chr12_-_76462713 | 0.06 |
ENST00000552056.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr3_+_88199099 | 0.06 |
ENST00000486971.1 |
C3orf38 |
chromosome 3 open reading frame 38 |
| chr4_+_1385340 | 0.06 |
ENST00000324803.4 |
CRIPAK |
cysteine-rich PAK1 inhibitor |
| chr19_-_59084922 | 0.06 |
ENST00000215057.2 ENST00000599369.1 |
MZF1 |
myeloid zinc finger 1 |
| chr18_-_37380230 | 0.06 |
ENST00000591629.1 |
LINC00669 |
long intergenic non-protein coding RNA 669 |
| chr1_-_35497506 | 0.06 |
ENST00000317538.5 ENST00000373340.2 ENST00000357182.4 |
ZMYM6 |
zinc finger, MYM-type 6 |
| chrX_-_102941596 | 0.06 |
ENST00000441076.2 ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr22_+_36649056 | 0.06 |
ENST00000397278.3 ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1 |
apolipoprotein L, 1 |
| chr11_+_6947647 | 0.06 |
ENST00000278319.5 |
ZNF215 |
zinc finger protein 215 |
| chr7_-_55606346 | 0.06 |
ENST00000545390.1 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr5_+_118690466 | 0.06 |
ENST00000503646.1 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
| chr17_+_3323862 | 0.06 |
ENST00000291231.1 |
OR3A3 |
olfactory receptor, family 3, subfamily A, member 3 |
| chr14_-_55658252 | 0.05 |
ENST00000395425.2 |
DLGAP5 |
discs, large (Drosophila) homolog-associated protein 5 |
| chr19_-_10491234 | 0.05 |
ENST00000524462.1 ENST00000531836.1 ENST00000525621.1 |
TYK2 |
tyrosine kinase 2 |
| chr11_-_18956556 | 0.05 |
ENST00000302797.3 |
MRGPRX1 |
MAS-related GPR, member X1 |
| chr17_+_41857793 | 0.05 |
ENST00000449302.3 |
C17orf105 |
chromosome 17 open reading frame 105 |
| chr3_-_139199565 | 0.05 |
ENST00000511956.1 |
RBP2 |
retinol binding protein 2, cellular |
| chr8_+_54764346 | 0.05 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chr15_-_99789736 | 0.05 |
ENST00000560235.1 ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr19_+_45418067 | 0.05 |
ENST00000589078.1 ENST00000586638.1 |
APOC1 |
apolipoprotein C-I |
| chr7_-_64023441 | 0.05 |
ENST00000309683.6 |
ZNF680 |
zinc finger protein 680 |
| chr4_+_71587669 | 0.05 |
ENST00000381006.3 ENST00000226328.4 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr4_+_76649797 | 0.05 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr16_+_446713 | 0.05 |
ENST00000397722.1 ENST00000454619.1 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
| chrM_+_7586 | 0.05 |
ENST00000361739.1 |
MT-CO2 |
mitochondrially encoded cytochrome c oxidase II |
| chr3_+_39424828 | 0.05 |
ENST00000273158.4 ENST00000431510.1 |
SLC25A38 |
solute carrier family 25, member 38 |
| chr12_-_75603482 | 0.05 |
ENST00000341669.3 ENST00000298972.1 ENST00000350228.2 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr19_-_35719609 | 0.05 |
ENST00000324675.3 |
FAM187B |
family with sequence similarity 187, member B |
| chr3_-_121468513 | 0.04 |
ENST00000494517.1 ENST00000393667.3 |
GOLGB1 |
golgin B1 |
| chr12_-_6579833 | 0.04 |
ENST00000396308.3 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr17_+_77030267 | 0.04 |
ENST00000581774.1 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr19_-_21950362 | 0.04 |
ENST00000358296.6 |
ZNF100 |
zinc finger protein 100 |
| chrM_+_5824 | 0.04 |
ENST00000361624.2 |
MT-CO1 |
mitochondrially encoded cytochrome c oxidase I |
| chr8_-_145652336 | 0.04 |
ENST00000529182.1 ENST00000526054.1 |
VPS28 |
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr3_-_121468602 | 0.04 |
ENST00000340645.5 |
GOLGB1 |
golgin B1 |
| chr12_-_11287243 | 0.04 |
ENST00000539585.1 |
TAS2R30 |
taste receptor, type 2, member 30 |
| chr19_+_45417921 | 0.04 |
ENST00000252491.4 ENST00000592885.1 ENST00000589781.1 |
APOC1 |
apolipoprotein C-I |
| chr3_-_133648656 | 0.04 |
ENST00000408895.2 |
C3orf36 |
chromosome 3 open reading frame 36 |
| chr7_-_137028498 | 0.04 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr1_+_100810575 | 0.04 |
ENST00000542213.1 |
CDC14A |
cell division cycle 14A |
| chr16_+_56672571 | 0.04 |
ENST00000290705.8 |
MT1A |
metallothionein 1A |
| chr2_-_136633940 | 0.04 |
ENST00000264156.2 |
MCM6 |
minichromosome maintenance complex component 6 |
| chr12_+_12223867 | 0.03 |
ENST00000308721.5 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
| chr16_+_50059125 | 0.03 |
ENST00000427478.2 |
CNEP1R1 |
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr4_+_71588372 | 0.03 |
ENST00000536664.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr12_+_12202774 | 0.03 |
ENST00000589718.1 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
| chr2_+_38177575 | 0.03 |
ENST00000407257.1 ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2 |
regulator of microtubule dynamics 2 |
| chr1_+_207669573 | 0.03 |
ENST00000400960.2 ENST00000534202.1 |
CR1 |
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr12_-_59175485 | 0.03 |
ENST00000550678.1 ENST00000552201.1 |
RP11-767I20.1 |
RP11-767I20.1 |
| chr12_+_12202785 | 0.03 |
ENST00000586576.1 ENST00000464885.2 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
| chr7_-_15601595 | 0.03 |
ENST00000342526.3 |
AGMO |
alkylglycerol monooxygenase |
| chr1_+_47603109 | 0.03 |
ENST00000371890.3 ENST00000294337.3 ENST00000371891.3 |
CYP4A22 |
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr17_-_42019836 | 0.03 |
ENST00000225992.3 |
PPY |
pancreatic polypeptide |
| chr7_-_137028534 | 0.03 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr6_+_118869452 | 0.03 |
ENST00000357525.5 |
PLN |
phospholamban |
| chr12_-_11463353 | 0.03 |
ENST00000279575.1 ENST00000535904.1 ENST00000445719.2 |
PRB4 |
proline-rich protein BstNI subfamily 4 |
| chrX_-_139866723 | 0.03 |
ENST00000370532.2 |
CDR1 |
cerebellar degeneration-related protein 1, 34kDa |
| chr20_+_21284416 | 0.02 |
ENST00000539513.1 |
XRN2 |
5'-3' exoribonuclease 2 |
| chr22_+_42017987 | 0.02 |
ENST00000405506.1 |
XRCC6 |
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr5_+_141488070 | 0.02 |
ENST00000253814.4 |
NDFIP1 |
Nedd4 family interacting protein 1 |
| chr1_+_36038971 | 0.02 |
ENST00000373235.3 |
TFAP2E |
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
| chr3_+_193965426 | 0.02 |
ENST00000455821.1 |
RP11-513G11.3 |
RP11-513G11.3 |
| chr21_-_44495919 | 0.02 |
ENST00000398158.1 |
CBS |
cystathionine-beta-synthase |
| chr14_-_46185155 | 0.02 |
ENST00000555442.1 |
RP11-369C8.1 |
RP11-369C8.1 |
| chrX_+_70503433 | 0.02 |
ENST00000276079.8 ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO |
non-POU domain containing, octamer-binding |
| chr4_-_17513851 | 0.02 |
ENST00000281243.5 |
QDPR |
quinoid dihydropteridine reductase |
| chr11_-_327537 | 0.02 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr14_+_101123580 | 0.02 |
ENST00000556697.1 ENST00000360899.2 ENST00000553623.1 |
LINC00523 |
long intergenic non-protein coding RNA 523 |
| chr1_+_174670143 | 0.02 |
ENST00000367687.1 ENST00000347255.2 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr2_-_127977654 | 0.02 |
ENST00000409327.1 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr6_+_116937636 | 0.02 |
ENST00000368581.4 ENST00000229554.5 ENST00000368580.4 |
RSPH4A |
radial spoke head 4 homolog A (Chlamydomonas) |
| chr22_+_25202232 | 0.02 |
ENST00000400358.4 ENST00000400359.4 |
SGSM1 |
small G protein signaling modulator 1 |
| chr16_+_72088376 | 0.02 |
ENST00000570083.1 ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP HPR |
haptoglobin haptoglobin-related protein |
| chr6_+_26104104 | 0.02 |
ENST00000377803.2 |
HIST1H4C |
histone cluster 1, H4c |
| chr1_-_198990166 | 0.02 |
ENST00000427439.1 |
RP11-16L9.3 |
RP11-16L9.3 |
| chr12_+_12224331 | 0.02 |
ENST00000396367.1 ENST00000266434.4 ENST00000396369.1 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
| chr10_-_46030787 | 0.01 |
ENST00000395769.2 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr16_-_28503327 | 0.01 |
ENST00000535392.1 ENST00000395653.4 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
| chr20_+_54823788 | 0.01 |
ENST00000243911.2 |
MC3R |
melanocortin 3 receptor |
| chr19_+_45417812 | 0.01 |
ENST00000592535.1 |
APOC1 |
apolipoprotein C-I |
| chr14_-_106668095 | 0.00 |
ENST00000390606.2 |
IGHV3-20 |
immunoglobulin heavy variable 3-20 |
| chr21_-_44495964 | 0.00 |
ENST00000398168.1 ENST00000398165.3 |
CBS |
cystathionine-beta-synthase |
| chr1_-_52344471 | 0.00 |
ENST00000352171.7 ENST00000354831.7 |
NRD1 |
nardilysin (N-arginine dibasic convertase) |
| chr2_-_198540719 | 0.00 |
ENST00000295049.4 |
RFTN2 |
raftlin family member 2 |
| chr16_+_75182376 | 0.00 |
ENST00000570010.1 ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1 |
ZFP1 zinc finger protein |
| chr10_+_82300575 | 0.00 |
ENST00000313455.4 |
SH2D4B |
SH2 domain containing 4B |
| chr14_-_75330537 | 0.00 |
ENST00000556084.2 ENST00000556489.2 ENST00000445876.1 |
PROX2 |
prospero homeobox 2 |
| chr2_-_89568263 | 0.00 |
ENST00000473726.1 |
IGKV1-33 |
immunoglobulin kappa variable 1-33 |
| chr1_-_161208013 | 0.00 |
ENST00000515452.1 ENST00000367983.4 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_154600421 | 0.00 |
ENST00000368471.3 ENST00000292205.5 |
ADAR |
adenosine deaminase, RNA-specific |
| chr17_+_34171081 | 0.00 |
ENST00000585577.1 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.4 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.3 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.5 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 1.0 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.6 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.4 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.9 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 1.0 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.1 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.0 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.0 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.0 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 2.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.1 | 0.6 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.4 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.0 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |