Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for ALX1_ARX
Z-value: 0.29
Transcription factors associated with ALX1_ARX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
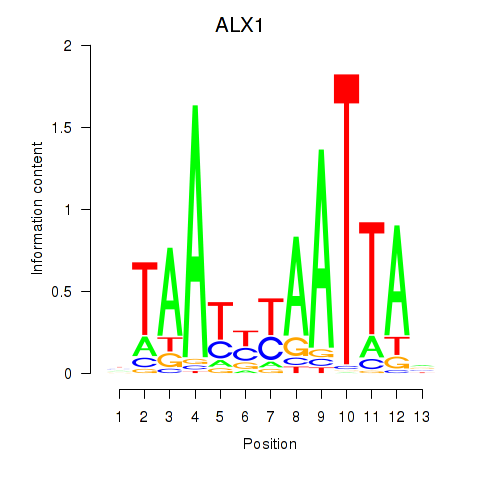
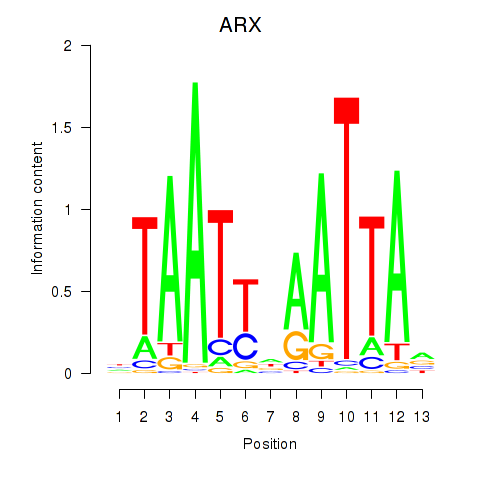
ALX1
|
ENSG00000180318.3 | ALX1 |
|
ARX
|
ENSG00000004848.6 | ARX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARX | hg19_v2_chrX_-_25034065_25034088 | -0.33 | 4.3e-01 | Click! |
| ALX1 | hg19_v2_chr12_+_85673868_85673885 | 0.09 | 8.3e-01 | Click! |
Activity profile of ALX1_ARX motif
Sorted Z-values of ALX1_ARX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX1_ARX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_69865866 | 0.75 |
ENST00000354393.2 |
MYPN |
myopalladin |
| chr1_-_92371839 | 0.52 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr12_-_91546926 | 0.46 |
ENST00000550758.1 |
DCN |
decorin |
| chr2_-_145278475 | 0.34 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr3_-_195310802 | 0.29 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr1_+_183774240 | 0.29 |
ENST00000360851.3 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr5_-_111312622 | 0.22 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr2_+_152214098 | 0.19 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr1_-_232651312 | 0.17 |
ENST00000262861.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr6_-_154751629 | 0.16 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr20_+_31823792 | 0.15 |
ENST00000375413.4 ENST00000354297.4 ENST00000375422.2 |
BPIFA1 |
BPI fold containing family A, member 1 |
| chr3_+_12329358 | 0.15 |
ENST00000309576.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr12_-_52946923 | 0.14 |
ENST00000267119.5 |
KRT71 |
keratin 71 |
| chr3_+_12329397 | 0.14 |
ENST00000397015.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr11_-_96076334 | 0.14 |
ENST00000524717.1 |
MAML2 |
mastermind-like 2 (Drosophila) |
| chrX_+_77166172 | 0.13 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr8_-_122653630 | 0.13 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr12_+_59989918 | 0.13 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr3_-_164914640 | 0.12 |
ENST00000241274.3 |
SLITRK3 |
SLIT and NTRK-like family, member 3 |
| chr7_+_90338712 | 0.11 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr12_+_18414446 | 0.11 |
ENST00000433979.1 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr10_+_89420706 | 0.11 |
ENST00000427144.2 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr6_+_34204642 | 0.11 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr16_+_53133070 | 0.11 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr17_+_39240459 | 0.10 |
ENST00000391417.4 |
KRTAP4-7 |
keratin associated protein 4-7 |
| chr2_-_145275228 | 0.10 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr11_-_82708435 | 0.10 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr3_-_157824292 | 0.09 |
ENST00000483851.2 |
SHOX2 |
short stature homeobox 2 |
| chr8_+_38677850 | 0.08 |
ENST00000518809.1 ENST00000520611.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chrX_-_15332665 | 0.08 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr15_-_52970820 | 0.08 |
ENST00000261844.7 ENST00000399202.4 ENST00000562135.1 |
FAM214A |
family with sequence similarity 214, member A |
| chr12_-_92539614 | 0.08 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr5_+_140762268 | 0.07 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr19_+_50016610 | 0.07 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr8_-_57123815 | 0.07 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr1_+_206557366 | 0.07 |
ENST00000414007.1 ENST00000419187.2 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
| chr10_-_31146615 | 0.07 |
ENST00000444692.2 |
ZNF438 |
zinc finger protein 438 |
| chr15_+_44719970 | 0.06 |
ENST00000558966.1 |
CTDSPL2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr5_-_160279207 | 0.06 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr14_+_95078714 | 0.06 |
ENST00000393078.3 ENST00000393080.4 ENST00000467132.1 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr8_-_26724784 | 0.06 |
ENST00000380573.3 |
ADRA1A |
adrenoceptor alpha 1A |
| chr3_+_111393501 | 0.06 |
ENST00000393934.3 |
PLCXD2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr2_-_145277882 | 0.06 |
ENST00000465070.1 ENST00000444559.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr17_+_44588877 | 0.06 |
ENST00000576629.1 |
LRRC37A2 |
leucine rich repeat containing 37, member A2 |
| chr4_-_83769996 | 0.06 |
ENST00000511338.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr17_-_5321549 | 0.05 |
ENST00000572809.1 |
NUP88 |
nucleoporin 88kDa |
| chr3_+_138340049 | 0.05 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr5_+_137203465 | 0.05 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr18_+_66465302 | 0.05 |
ENST00000360242.5 ENST00000358653.5 |
CCDC102B |
coiled-coil domain containing 102B |
| chr10_-_29923893 | 0.05 |
ENST00000355867.4 |
SVIL |
supervillin |
| chr1_+_84630645 | 0.05 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_-_12946025 | 0.05 |
ENST00000235349.5 |
PRAMEF4 |
PRAME family member 4 |
| chr15_-_26874230 | 0.05 |
ENST00000400188.3 |
GABRB3 |
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr9_-_95298314 | 0.05 |
ENST00000344604.5 ENST00000375540.1 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
| chr5_+_140743859 | 0.05 |
ENST00000518069.1 |
PCDHGA5 |
protocadherin gamma subfamily A, 5 |
| chr13_-_81801115 | 0.05 |
ENST00000567258.1 |
LINC00564 |
long intergenic non-protein coding RNA 564 |
| chr7_-_140482926 | 0.05 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr7_+_138915102 | 0.05 |
ENST00000486663.1 |
UBN2 |
ubinuclein 2 |
| chr10_+_134150835 | 0.05 |
ENST00000432555.2 |
LRRC27 |
leucine rich repeat containing 27 |
| chr6_+_31895254 | 0.05 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chr5_+_59783540 | 0.05 |
ENST00000515734.2 |
PART1 |
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr2_+_179318295 | 0.05 |
ENST00000442710.1 |
DFNB59 |
deafness, autosomal recessive 59 |
| chr2_+_161993465 | 0.05 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr3_+_40518599 | 0.04 |
ENST00000314686.5 ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619 |
zinc finger protein 619 |
| chr14_+_57671888 | 0.04 |
ENST00000391612.1 |
AL391152.1 |
AL391152.1 |
| chrX_+_36246735 | 0.04 |
ENST00000378653.3 |
CXorf30 |
chromosome X open reading frame 30 |
| chr2_+_65215604 | 0.04 |
ENST00000531327.1 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr4_-_119759795 | 0.04 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr13_+_53602894 | 0.04 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr1_+_44679159 | 0.04 |
ENST00000315913.5 ENST00000372289.2 |
DMAP1 |
DNA methyltransferase 1 associated protein 1 |
| chr2_+_101437487 | 0.04 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr1_+_158978768 | 0.04 |
ENST00000447473.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr1_+_44679113 | 0.04 |
ENST00000361745.6 ENST00000446292.1 ENST00000440641.1 ENST00000436069.1 ENST00000437511.1 |
DMAP1 |
DNA methyltransferase 1 associated protein 1 |
| chr2_+_161993412 | 0.04 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr3_-_124774802 | 0.04 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr17_-_71228357 | 0.04 |
ENST00000583024.1 ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A |
family with sequence similarity 104, member A |
| chrX_+_36254051 | 0.04 |
ENST00000378657.4 |
CXorf30 |
chromosome X open reading frame 30 |
| chr11_+_120973375 | 0.04 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr12_-_118796910 | 0.04 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr7_+_148287657 | 0.04 |
ENST00000307003.2 |
C7orf33 |
chromosome 7 open reading frame 33 |
| chr1_+_154401791 | 0.04 |
ENST00000476006.1 |
IL6R |
interleukin 6 receptor |
| chr8_+_11961898 | 0.03 |
ENST00000400085.3 |
ZNF705D |
zinc finger protein 705D |
| chrX_-_48858667 | 0.03 |
ENST00000376423.4 ENST00000376441.1 |
GRIPAP1 |
GRIP1 associated protein 1 |
| chr12_+_12878829 | 0.03 |
ENST00000326765.6 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr12_-_53601055 | 0.03 |
ENST00000552972.1 ENST00000422257.3 ENST00000267082.5 |
ITGB7 |
integrin, beta 7 |
| chr15_+_51669444 | 0.03 |
ENST00000396399.2 |
GLDN |
gliomedin |
| chr3_+_138340067 | 0.03 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chrX_-_48858630 | 0.03 |
ENST00000376425.3 ENST00000376444.3 |
GRIPAP1 |
GRIP1 associated protein 1 |
| chr1_+_52682052 | 0.03 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr4_+_113568207 | 0.03 |
ENST00000511529.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr12_-_53601000 | 0.03 |
ENST00000338737.4 ENST00000549086.2 |
ITGB7 |
integrin, beta 7 |
| chr8_-_62602327 | 0.03 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr4_+_74347400 | 0.03 |
ENST00000226355.3 |
AFM |
afamin |
| chr3_-_47934234 | 0.03 |
ENST00000420772.2 |
MAP4 |
microtubule-associated protein 4 |
| chr7_+_90339169 | 0.03 |
ENST00000436577.2 |
CDK14 |
cyclin-dependent kinase 14 |
| chr19_-_6393216 | 0.03 |
ENST00000595047.1 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
| chrX_-_19817869 | 0.03 |
ENST00000379698.4 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr6_-_111804905 | 0.03 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr7_-_14880892 | 0.03 |
ENST00000406247.3 ENST00000399322.3 ENST00000258767.5 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr5_+_68860949 | 0.03 |
ENST00000507595.1 |
GTF2H2C |
general transcription factor IIH, polypeptide 2C |
| chr4_-_83765613 | 0.03 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr2_+_54683419 | 0.03 |
ENST00000356805.4 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr5_+_140514782 | 0.03 |
ENST00000231134.5 |
PCDHB5 |
protocadherin beta 5 |
| chr17_-_39623681 | 0.03 |
ENST00000225899.3 |
KRT32 |
keratin 32 |
| chr17_+_71228793 | 0.03 |
ENST00000426147.2 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr5_+_140079919 | 0.03 |
ENST00000274712.3 |
ZMAT2 |
zinc finger, matrin-type 2 |
| chr14_-_81425828 | 0.03 |
ENST00000555529.1 ENST00000556042.1 ENST00000556981.1 |
CEP128 |
centrosomal protein 128kDa |
| chr7_+_55980331 | 0.03 |
ENST00000429591.2 |
ZNF713 |
zinc finger protein 713 |
| chrX_+_16668278 | 0.02 |
ENST00000380200.3 |
S100G |
S100 calcium binding protein G |
| chr14_-_101036119 | 0.02 |
ENST00000355173.2 |
BEGAIN |
brain-enriched guanylate kinase-associated |
| chr15_-_75748115 | 0.02 |
ENST00000360439.4 |
SIN3A |
SIN3 transcription regulator family member A |
| chr17_+_71228537 | 0.02 |
ENST00000577615.1 ENST00000585109.1 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr21_+_48055527 | 0.02 |
ENST00000397638.2 ENST00000458387.2 ENST00000451211.2 ENST00000291705.6 ENST00000397637.1 ENST00000334494.4 ENST00000397628.1 ENST00000440086.1 |
PRMT2 |
protein arginine methyltransferase 2 |
| chr6_-_107235331 | 0.02 |
ENST00000433965.1 ENST00000430094.1 |
RP1-60O19.1 |
RP1-60O19.1 |
| chr1_+_192127578 | 0.02 |
ENST00000367460.3 |
RGS18 |
regulator of G-protein signaling 18 |
| chr2_+_228678550 | 0.02 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr10_+_53806501 | 0.02 |
ENST00000373975.2 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr10_+_35464513 | 0.02 |
ENST00000494479.1 ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM |
cAMP responsive element modulator |
| chr19_-_54619006 | 0.02 |
ENST00000391759.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chrX_+_38211777 | 0.02 |
ENST00000039007.4 |
OTC |
ornithine carbamoyltransferase |
| chr4_+_88754113 | 0.02 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr18_-_21891460 | 0.02 |
ENST00000357041.4 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr3_-_186262166 | 0.02 |
ENST00000307944.5 |
CRYGS |
crystallin, gamma S |
| chr5_-_39270725 | 0.02 |
ENST00000512138.1 ENST00000512982.1 ENST00000540520.1 |
FYB |
FYN binding protein |
| chr5_+_38148582 | 0.02 |
ENST00000508853.1 |
CTD-2207A17.1 |
CTD-2207A17.1 |
| chr4_+_76649797 | 0.02 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr21_+_17442799 | 0.02 |
ENST00000602580.1 ENST00000458468.1 ENST00000602935.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr8_+_7353368 | 0.02 |
ENST00000355602.2 |
DEFB107B |
defensin, beta 107B |
| chr5_+_59783941 | 0.02 |
ENST00000506884.1 ENST00000504876.2 |
PART1 |
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr1_+_186265399 | 0.01 |
ENST00000367486.3 ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4 |
proteoglycan 4 |
| chr11_+_62496114 | 0.01 |
ENST00000532583.1 |
TTC9C |
tetratricopeptide repeat domain 9C |
| chr5_+_133562095 | 0.01 |
ENST00000602919.1 |
CTD-2410N18.3 |
CTD-2410N18.3 |
| chr15_+_43477580 | 0.01 |
ENST00000356633.5 |
CCNDBP1 |
cyclin D-type binding-protein 1 |
| chr6_-_76072719 | 0.01 |
ENST00000370020.1 |
FILIP1 |
filamin A interacting protein 1 |
| chr2_-_70529180 | 0.01 |
ENST00000450256.1 ENST00000037869.3 |
FAM136A |
family with sequence similarity 136, member A |
| chr17_-_3030875 | 0.01 |
ENST00000328890.2 |
OR1G1 |
olfactory receptor, family 1, subfamily G, member 1 |
| chr15_+_66874502 | 0.01 |
ENST00000558797.1 |
RP11-321F6.1 |
HCG2003567; Uncharacterized protein |
| chr3_+_159943362 | 0.01 |
ENST00000326474.3 |
C3orf80 |
chromosome 3 open reading frame 80 |
| chr5_+_137203557 | 0.01 |
ENST00000515645.1 |
MYOT |
myotilin |
| chr14_+_24099318 | 0.01 |
ENST00000432832.2 |
DHRS2 |
dehydrogenase/reductase (SDR family) member 2 |
| chr17_+_75181292 | 0.01 |
ENST00000431431.2 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr16_+_1728257 | 0.01 |
ENST00000248098.3 ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L |
hematological and neurological expressed 1-like |
| chr16_+_68119247 | 0.01 |
ENST00000575270.1 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr17_+_71228740 | 0.01 |
ENST00000268942.8 ENST00000359042.2 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr12_-_10151773 | 0.01 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr15_+_57511609 | 0.01 |
ENST00000543579.1 ENST00000537840.1 ENST00000343827.3 |
TCF12 |
transcription factor 12 |
| chr6_-_150067696 | 0.01 |
ENST00000340413.2 ENST00000367403.3 |
NUP43 |
nucleoporin 43kDa |
| chr1_-_151148492 | 0.01 |
ENST00000295314.4 |
TMOD4 |
tropomodulin 4 (muscle) |
| chr3_-_12587055 | 0.01 |
ENST00000564146.3 |
C3orf83 |
chromosome 3 open reading frame 83 |
| chr8_+_77318769 | 0.01 |
ENST00000518732.1 |
RP11-706J10.1 |
long intergenic non-protein coding RNA 1111 |
| chr11_-_111649015 | 0.01 |
ENST00000529841.1 |
RP11-108O10.2 |
RP11-108O10.2 |
| chr8_-_57472154 | 0.01 |
ENST00000499425.1 ENST00000523664.1 ENST00000518943.1 ENST00000524338.1 |
LINC00968 |
long intergenic non-protein coding RNA 968 |
| chr9_+_44867571 | 0.01 |
ENST00000377548.2 |
RP11-160N1.10 |
RP11-160N1.10 |
| chr19_-_6393465 | 0.01 |
ENST00000394456.5 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
| chrX_-_138724994 | 0.01 |
ENST00000536274.1 |
MCF2 |
MCF.2 cell line derived transforming sequence |
| chr4_-_100356291 | 0.01 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_133055815 | 0.01 |
ENST00000509351.1 ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3 |
vanin 3 |
| chrX_+_35937843 | 0.01 |
ENST00000297866.5 |
CXorf22 |
chromosome X open reading frame 22 |
| chr2_+_74685413 | 0.01 |
ENST00000233615.2 |
WBP1 |
WW domain binding protein 1 |
| chr3_+_155860751 | 0.01 |
ENST00000471742.1 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr6_-_133055896 | 0.01 |
ENST00000367927.5 ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3 |
vanin 3 |
| chr7_-_7575477 | 0.01 |
ENST00000399429.3 |
COL28A1 |
collagen, type XXVIII, alpha 1 |
| chr11_+_95523621 | 0.01 |
ENST00000325542.5 ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57 |
centrosomal protein 57kDa |
| chr11_-_58499434 | 0.01 |
ENST00000344743.3 ENST00000278400.3 |
GLYAT |
glycine-N-acyltransferase |
| chr8_-_61880248 | 0.01 |
ENST00000525556.1 |
AC022182.3 |
AC022182.3 |
| chr12_-_57873631 | 0.01 |
ENST00000393791.3 ENST00000356411.2 ENST00000552249.1 |
ARHGAP9 |
Rho GTPase activating protein 9 |
| chr12_+_122241928 | 0.01 |
ENST00000604567.1 ENST00000542440.1 |
SETD1B |
SET domain containing 1B |
| chr4_-_89951028 | 0.01 |
ENST00000506913.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr2_+_119699742 | 0.01 |
ENST00000327097.4 |
MARCO |
macrophage receptor with collagenous structure |
| chrM_+_7586 | 0.00 |
ENST00000361739.1 |
MT-CO2 |
mitochondrially encoded cytochrome c oxidase II |
| chr9_+_77230499 | 0.00 |
ENST00000396204.2 |
RORB |
RAR-related orphan receptor B |
| chr1_-_151148442 | 0.00 |
ENST00000441701.1 ENST00000416280.2 |
TMOD4 |
tropomodulin 4 (muscle) |
| chr1_-_154600421 | 0.00 |
ENST00000368471.3 ENST00000292205.5 |
ADAR |
adenosine deaminase, RNA-specific |
| chr1_-_13390765 | 0.00 |
ENST00000357367.2 |
PRAMEF8 |
PRAME family member 8 |
| chr19_-_58485895 | 0.00 |
ENST00000314391.3 |
C19orf18 |
chromosome 19 open reading frame 18 |
| chr6_-_41039567 | 0.00 |
ENST00000468811.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr16_+_68119324 | 0.00 |
ENST00000349223.5 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr15_+_44092784 | 0.00 |
ENST00000458412.1 |
HYPK |
huntingtin interacting protein K |
| chr2_+_36923830 | 0.00 |
ENST00000379242.3 ENST00000389975.3 |
VIT |
vitrin |
| chr3_+_151591422 | 0.00 |
ENST00000362032.5 |
SUCNR1 |
succinate receptor 1 |
| chr1_-_110155671 | 0.00 |
ENST00000351050.3 |
GNAT2 |
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr1_+_104615595 | 0.00 |
ENST00000418362.1 |
RP11-364B6.1 |
RP11-364B6.1 |
| chr3_+_140660743 | 0.00 |
ENST00000453248.2 |
SLC25A36 |
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr14_+_38264418 | 0.00 |
ENST00000267368.7 ENST00000382320.3 |
TTC6 |
tetratricopeptide repeat domain 6 |
| chr2_+_185463093 | 0.00 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr3_+_39424828 | 0.00 |
ENST00000273158.4 ENST00000431510.1 |
SLC25A38 |
solute carrier family 25, member 38 |
| chr3_-_101039402 | 0.00 |
ENST00000193391.7 |
IMPG2 |
interphotoreceptor matrix proteoglycan 2 |
| chr4_+_95128748 | 0.00 |
ENST00000359052.4 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr11_+_18433840 | 0.00 |
ENST00000541669.1 ENST00000280704.4 |
LDHC |
lactate dehydrogenase C |
| chr16_+_68119440 | 0.00 |
ENST00000346183.3 ENST00000329524.4 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:0003099 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |