Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
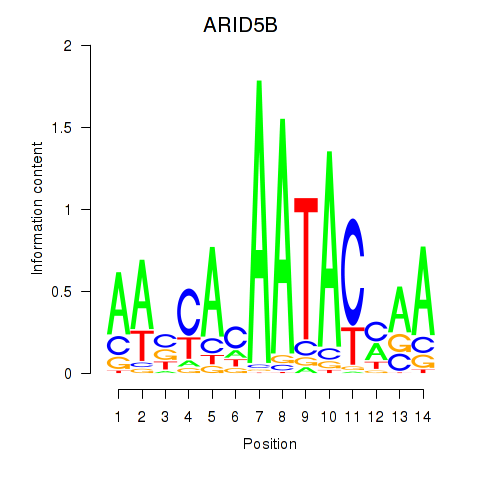
Results for ARID5B
Z-value: 0.33
Transcription factors associated with ARID5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5B
|
ENSG00000150347.10 | ARID5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5B | hg19_v2_chr10_+_63661053_63661079 | 0.47 | 2.4e-01 | Click! |
Activity profile of ARID5B motif
Sorted Z-values of ARID5B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5B
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91539918 | 0.37 |
ENST00000548218.1 |
DCN |
decorin |
| chr5_+_92919043 | 0.33 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr7_-_56119156 | 0.31 |
ENST00000421312.1 ENST00000416592.1 |
PSPH |
phosphoserine phosphatase |
| chr5_+_32788945 | 0.30 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr3_+_99357319 | 0.30 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr4_-_159094194 | 0.27 |
ENST00000592057.1 ENST00000585682.1 ENST00000393807.5 |
FAM198B |
family with sequence similarity 198, member B |
| chr12_-_15038779 | 0.24 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr4_-_57524061 | 0.22 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chrX_+_102585124 | 0.21 |
ENST00000332431.4 ENST00000372666.1 |
TCEAL7 |
transcription elongation factor A (SII)-like 7 |
| chr1_+_151739131 | 0.21 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chrX_+_86772787 | 0.19 |
ENST00000373114.4 |
KLHL4 |
kelch-like family member 4 |
| chr1_-_168698433 | 0.19 |
ENST00000367817.3 |
DPT |
dermatopontin |
| chr22_-_22090043 | 0.18 |
ENST00000403503.1 |
YPEL1 |
yippee-like 1 (Drosophila) |
| chr14_+_22458631 | 0.18 |
ENST00000390444.1 |
TRAV16 |
T cell receptor alpha variable 16 |
| chr9_-_123812542 | 0.17 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr7_-_56119238 | 0.17 |
ENST00000275605.3 ENST00000395471.3 |
PSPH |
phosphoserine phosphatase |
| chr19_-_3029011 | 0.16 |
ENST00000590536.1 ENST00000587137.1 ENST00000455444.2 ENST00000262953.6 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr11_-_111794446 | 0.15 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr1_+_61330931 | 0.14 |
ENST00000371191.1 |
NFIA |
nuclear factor I/A |
| chr6_+_32812568 | 0.14 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr4_+_71458012 | 0.14 |
ENST00000449493.2 |
AMBN |
ameloblastin (enamel matrix protein) |
| chr3_-_114790179 | 0.13 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr9_-_116065551 | 0.12 |
ENST00000297894.5 |
RNF183 |
ring finger protein 183 |
| chr12_+_8309630 | 0.12 |
ENST00000396570.3 |
ZNF705A |
zinc finger protein 705A |
| chrX_-_49089771 | 0.12 |
ENST00000376251.1 ENST00000323022.5 ENST00000376265.2 |
CACNA1F |
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr15_+_25068773 | 0.12 |
ENST00000400100.1 ENST00000400098.1 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr1_-_160231451 | 0.11 |
ENST00000495887.1 |
DCAF8 |
DDB1 and CUL4 associated factor 8 |
| chr8_-_116504448 | 0.11 |
ENST00000518018.1 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr8_-_101321584 | 0.11 |
ENST00000523167.1 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr19_+_17326141 | 0.11 |
ENST00000445667.2 ENST00000263897.5 |
USE1 |
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr19_+_35849362 | 0.11 |
ENST00000327809.4 |
FFAR3 |
free fatty acid receptor 3 |
| chr21_+_30502806 | 0.11 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr22_+_42086547 | 0.10 |
ENST00000402966.1 |
C22orf46 |
chromosome 22 open reading frame 46 |
| chr10_+_126150369 | 0.10 |
ENST00000392757.4 ENST00000368842.5 ENST00000368839.1 |
LHPP |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr1_-_144995074 | 0.10 |
ENST00000534536.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr1_+_171107241 | 0.10 |
ENST00000236166.3 |
FMO6P |
flavin containing monooxygenase 6 pseudogene |
| chr1_-_144994909 | 0.10 |
ENST00000369347.4 ENST00000369354.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr1_-_153085984 | 0.10 |
ENST00000468739.1 |
SPRR2F |
small proline-rich protein 2F |
| chr15_+_51669444 | 0.10 |
ENST00000396399.2 |
GLDN |
gliomedin |
| chr1_+_6615241 | 0.09 |
ENST00000333172.6 ENST00000328191.4 ENST00000351136.3 |
TAS1R1 |
taste receptor, type 1, member 1 |
| chr18_-_25616519 | 0.09 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr8_-_72268889 | 0.09 |
ENST00000388742.4 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr5_-_64920115 | 0.09 |
ENST00000381018.3 ENST00000274327.7 |
TRIM23 |
tripartite motif containing 23 |
| chr2_+_89999259 | 0.08 |
ENST00000558026.1 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chrX_+_73164167 | 0.08 |
ENST00000414209.1 ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX |
JPX transcript, XIST activator (non-protein coding) |
| chr4_+_189060573 | 0.08 |
ENST00000332517.3 |
TRIML1 |
tripartite motif family-like 1 |
| chr12_+_79258547 | 0.08 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr1_+_150266256 | 0.08 |
ENST00000309092.7 ENST00000369084.5 |
MRPS21 |
mitochondrial ribosomal protein S21 |
| chrX_-_57164058 | 0.07 |
ENST00000374906.3 |
SPIN2A |
spindlin family, member 2A |
| chr10_+_53806501 | 0.07 |
ENST00000373975.2 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr1_+_158975744 | 0.07 |
ENST00000426592.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr8_-_72268968 | 0.07 |
ENST00000388740.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr18_+_32556892 | 0.07 |
ENST00000591734.1 ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr14_-_105708942 | 0.07 |
ENST00000549655.1 |
BRF1 |
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr22_-_29107919 | 0.07 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr6_+_80341000 | 0.06 |
ENST00000369838.4 |
SH3BGRL2 |
SH3 domain binding glutamic acid-rich protein like 2 |
| chr12_+_79258444 | 0.06 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr5_+_161495038 | 0.06 |
ENST00000393933.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chrX_-_101410762 | 0.06 |
ENST00000543160.1 ENST00000333643.3 |
BEX5 |
brain expressed, X-linked 5 |
| chr5_-_179047881 | 0.06 |
ENST00000521173.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr4_-_89978299 | 0.06 |
ENST00000511976.1 ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr17_-_76713100 | 0.06 |
ENST00000585509.1 |
CYTH1 |
cytohesin 1 |
| chr5_+_135496675 | 0.05 |
ENST00000507637.1 |
SMAD5 |
SMAD family member 5 |
| chr1_+_104159999 | 0.05 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr1_+_172628154 | 0.05 |
ENST00000340030.3 ENST00000367721.2 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
| chr11_-_102714534 | 0.05 |
ENST00000299855.5 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr5_+_118691706 | 0.05 |
ENST00000415806.2 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
| chr1_+_32608566 | 0.05 |
ENST00000545542.1 |
KPNA6 |
karyopherin alpha 6 (importin alpha 7) |
| chr2_+_119699864 | 0.05 |
ENST00000541757.1 ENST00000412481.1 |
MARCO |
macrophage receptor with collagenous structure |
| chr1_-_43855479 | 0.05 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr11_-_74178582 | 0.05 |
ENST00000531854.1 ENST00000526855.1 ENST00000529425.1 ENST00000310128.4 ENST00000532569.1 |
KCNE3 |
potassium voltage-gated channel, Isk-related family, member 3 |
| chr8_-_145652336 | 0.05 |
ENST00000529182.1 ENST00000526054.1 |
VPS28 |
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr3_-_151102529 | 0.05 |
ENST00000302632.3 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
| chr12_-_56321649 | 0.04 |
ENST00000454792.2 ENST00000408946.2 |
WIBG |
within bgcn homolog (Drosophila) |
| chr6_-_15548591 | 0.04 |
ENST00000509674.1 |
DTNBP1 |
dystrobrevin binding protein 1 |
| chr3_+_196594727 | 0.04 |
ENST00000445299.2 ENST00000323460.5 ENST00000419026.1 |
SENP5 |
SUMO1/sentrin specific peptidase 5 |
| chr4_+_71588372 | 0.04 |
ENST00000536664.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr15_+_36994210 | 0.04 |
ENST00000562489.1 |
C15orf41 |
chromosome 15 open reading frame 41 |
| chr12_+_56390964 | 0.04 |
ENST00000356124.4 ENST00000266971.3 ENST00000394115.2 ENST00000547586.1 ENST00000552258.1 ENST00000548274.1 ENST00000546833.1 |
SUOX |
sulfite oxidase |
| chr8_-_66474884 | 0.04 |
ENST00000520902.1 |
CTD-3025N20.2 |
CTD-3025N20.2 |
| chr18_+_616711 | 0.04 |
ENST00000579494.1 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr1_-_144994840 | 0.04 |
ENST00000369351.3 ENST00000369349.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr3_-_185538849 | 0.03 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr2_-_40680578 | 0.03 |
ENST00000455476.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr6_+_29068386 | 0.03 |
ENST00000377171.3 |
OR2J1 |
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr7_+_149535455 | 0.03 |
ENST00000223210.4 ENST00000460379.1 |
ZNF862 |
zinc finger protein 862 |
| chr12_-_56321397 | 0.03 |
ENST00000557259.1 ENST00000549939.1 |
WIBG |
within bgcn homolog (Drosophila) |
| chr9_-_21202204 | 0.03 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr22_-_42086477 | 0.03 |
ENST00000402458.1 |
NHP2L1 |
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr11_-_78052923 | 0.03 |
ENST00000340149.2 |
GAB2 |
GRB2-associated binding protein 2 |
| chr11_+_33278811 | 0.03 |
ENST00000303296.4 ENST00000379016.3 |
HIPK3 |
homeodomain interacting protein kinase 3 |
| chrX_+_54466829 | 0.03 |
ENST00000375151.4 |
TSR2 |
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr2_-_158345462 | 0.03 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr18_+_9102592 | 0.03 |
ENST00000318388.6 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr3_-_15540055 | 0.03 |
ENST00000605797.1 ENST00000435459.2 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr9_+_77230499 | 0.03 |
ENST00000396204.2 |
RORB |
RAR-related orphan receptor B |
| chr17_-_21454898 | 0.03 |
ENST00000391411.5 ENST00000412778.3 |
C17orf51 |
chromosome 17 open reading frame 51 |
| chr8_+_55528627 | 0.03 |
ENST00000220676.1 |
RP1 |
retinitis pigmentosa 1 (autosomal dominant) |
| chr4_+_184020398 | 0.03 |
ENST00000403733.3 ENST00000378925.3 |
WWC2 |
WW and C2 domain containing 2 |
| chr2_-_74618964 | 0.03 |
ENST00000417090.1 ENST00000409868.1 |
DCTN1 |
dynactin 1 |
| chr7_+_153749732 | 0.03 |
ENST00000377770.3 |
DPP6 |
dipeptidyl-peptidase 6 |
| chr1_-_115292591 | 0.03 |
ENST00000438362.2 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chr1_-_40137710 | 0.03 |
ENST00000235628.1 |
NT5C1A |
5'-nucleotidase, cytosolic IA |
| chr5_+_175288631 | 0.03 |
ENST00000509837.1 |
CPLX2 |
complexin 2 |
| chr8_-_143957213 | 0.03 |
ENST00000519285.1 |
CYP11B1 |
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| chr6_+_132891461 | 0.03 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr11_+_101983176 | 0.03 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr7_-_115608304 | 0.02 |
ENST00000457268.1 |
TFEC |
transcription factor EC |
| chr1_-_114414316 | 0.02 |
ENST00000528414.1 ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22 |
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr14_-_69263043 | 0.02 |
ENST00000408913.2 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
| chr14_+_96968707 | 0.02 |
ENST00000216277.8 ENST00000557320.1 ENST00000557471.1 |
PAPOLA |
poly(A) polymerase alpha |
| chr2_-_74619152 | 0.02 |
ENST00000440727.1 ENST00000409240.1 |
DCTN1 |
dynactin 1 |
| chr14_-_24711806 | 0.02 |
ENST00000540705.1 ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr21_+_40177143 | 0.02 |
ENST00000360214.3 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chrX_+_140677562 | 0.02 |
ENST00000370518.3 |
SPANXA2 |
SPANX family, member A2 |
| chr1_+_104293028 | 0.02 |
ENST00000370079.3 |
AMY1C |
amylase, alpha 1C (salivary) |
| chr10_-_29923893 | 0.02 |
ENST00000355867.4 |
SVIL |
supervillin |
| chr4_+_187187098 | 0.02 |
ENST00000403665.2 ENST00000264692.4 |
F11 |
coagulation factor XI |
| chr14_-_65409502 | 0.02 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr4_-_69817481 | 0.02 |
ENST00000251566.4 |
UGT2A3 |
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chrX_+_140084756 | 0.02 |
ENST00000449283.1 |
SPANXB2 |
SPANX family, member B2 |
| chr1_-_43855444 | 0.02 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr12_-_91573249 | 0.02 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr14_-_24711865 | 0.02 |
ENST00000399423.4 ENST00000267415.7 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr6_-_30080863 | 0.02 |
ENST00000540829.1 |
TRIM31 |
tripartite motif containing 31 |
| chr6_+_122720681 | 0.02 |
ENST00000368455.4 ENST00000452194.1 |
HSF2 |
heat shock transcription factor 2 |
| chr7_+_153749471 | 0.02 |
ENST00000406326.1 |
DPP6 |
dipeptidyl-peptidase 6 |
| chr22_-_22090064 | 0.02 |
ENST00000339468.3 |
YPEL1 |
yippee-like 1 (Drosophila) |
| chr1_-_205391178 | 0.01 |
ENST00000367153.4 ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1 |
LEM domain containing 1 |
| chr17_+_37356586 | 0.01 |
ENST00000579260.1 ENST00000582193.1 |
RPL19 |
ribosomal protein L19 |
| chr12_+_93096619 | 0.01 |
ENST00000397833.3 |
C12orf74 |
chromosome 12 open reading frame 74 |
| chr19_+_9361606 | 0.01 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chr6_+_42896865 | 0.01 |
ENST00000372836.4 ENST00000394142.3 |
CNPY3 |
canopy FGF signaling regulator 3 |
| chr5_+_446253 | 0.01 |
ENST00000315013.5 |
EXOC3 |
exocyst complex component 3 |
| chr13_+_53216565 | 0.01 |
ENST00000357495.2 |
HNRNPA1L2 |
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr5_-_88179302 | 0.01 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr17_+_37356555 | 0.01 |
ENST00000579374.1 |
RPL19 |
ribosomal protein L19 |
| chr17_+_37356528 | 0.01 |
ENST00000225430.4 |
RPL19 |
ribosomal protein L19 |
| chr17_-_39341594 | 0.01 |
ENST00000398472.1 |
KRTAP4-1 |
keratin associated protein 4-1 |
| chr12_-_68553512 | 0.01 |
ENST00000229135.3 |
IFNG |
interferon, gamma |
| chr15_-_55563072 | 0.01 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr8_-_101322132 | 0.01 |
ENST00000523481.1 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr7_-_155604967 | 0.01 |
ENST00000297261.2 |
SHH |
sonic hedgehog |
| chr14_+_32798462 | 0.01 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr7_-_104909435 | 0.01 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr12_-_122879969 | 0.01 |
ENST00000540304.1 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
| chr3_+_102153859 | 0.01 |
ENST00000306176.1 ENST00000466937.1 |
ZPLD1 |
zona pellucida-like domain containing 1 |
| chr3_+_178276488 | 0.01 |
ENST00000432997.1 ENST00000455865.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr5_+_155753745 | 0.01 |
ENST00000435422.3 ENST00000337851.4 ENST00000447401.1 |
SGCD |
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr12_-_6809958 | 0.01 |
ENST00000320591.5 ENST00000534837.1 |
PIANP |
PILR alpha associated neural protein |
| chr19_+_57640011 | 0.01 |
ENST00000598197.1 |
USP29 |
ubiquitin specific peptidase 29 |
| chr11_-_104769141 | 0.01 |
ENST00000508062.1 ENST00000422698.2 |
CASP12 |
caspase 12 (gene/pseudogene) |
| chr1_+_228870824 | 0.01 |
ENST00000366691.3 |
RHOU |
ras homolog family member U |
| chr1_+_47264711 | 0.01 |
ENST00000371923.4 ENST00000271153.4 ENST00000371919.4 |
CYP4B1 |
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr11_-_73882057 | 0.01 |
ENST00000334126.7 ENST00000313663.7 |
C2CD3 |
C2 calcium-dependent domain containing 3 |
| chr10_-_69834973 | 0.01 |
ENST00000395187.2 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr1_-_76076793 | 0.00 |
ENST00000370859.3 |
SLC44A5 |
solute carrier family 44, member 5 |
| chr17_+_3379284 | 0.00 |
ENST00000263080.2 |
ASPA |
aspartoacylase |
| chr10_+_4828815 | 0.00 |
ENST00000533295.1 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
| chr12_+_120933904 | 0.00 |
ENST00000550178.1 ENST00000550845.1 ENST00000549989.1 ENST00000552870.1 |
DYNLL1 |
dynein, light chain, LC8-type 1 |
| chr6_+_30130969 | 0.00 |
ENST00000376694.4 |
TRIM15 |
tripartite motif containing 15 |
| chr2_+_181845843 | 0.00 |
ENST00000602710.1 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chrX_+_106769876 | 0.00 |
ENST00000439554.1 |
FRMPD3 |
FERM and PDZ domain containing 3 |
| chr1_+_150039369 | 0.00 |
ENST00000369130.3 ENST00000369128.5 |
VPS45 |
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr22_+_39916558 | 0.00 |
ENST00000337304.2 ENST00000396680.1 |
ATF4 |
activating transcription factor 4 |
| chr2_-_89327228 | 0.00 |
ENST00000483158.1 |
IGKV3-11 |
immunoglobulin kappa variable 3-11 |
| chr22_+_40573921 | 0.00 |
ENST00000454349.2 ENST00000335727.9 |
TNRC6B |
trinucleotide repeat containing 6B |
| chr15_+_69857515 | 0.00 |
ENST00000559477.1 |
RP11-279F6.1 |
RP11-279F6.1 |
| chr6_-_137494775 | 0.00 |
ENST00000349184.4 ENST00000296980.2 ENST00000339602.3 |
IL22RA2 |
interleukin 22 receptor, alpha 2 |
| chr2_-_231825668 | 0.00 |
ENST00000392039.2 |
GPR55 |
G protein-coupled receptor 55 |
| chr4_+_106631966 | 0.00 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr9_+_114287433 | 0.00 |
ENST00000358151.4 ENST00000355824.3 ENST00000374374.3 ENST00000309235.5 |
ZNF483 |
zinc finger protein 483 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.0 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |