Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for ATF6
Z-value: 1.59
Transcription factors associated with ATF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF6
|
ENSG00000118217.5 | ATF6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF6 | hg19_v2_chr1_+_161736072_161736093 | -0.51 | 2.0e-01 | Click! |
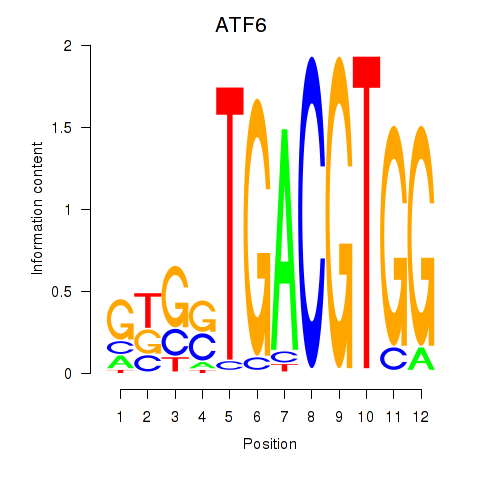
Activity profile of ATF6 motif
Sorted Z-values of ATF6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_45067659 | 2.47 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr4_+_145567173 | 2.31 |
ENST00000296575.3 |
HHIP |
hedgehog interacting protein |
| chr11_-_35547572 | 2.22 |
ENST00000378880.2 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr11_+_113930291 | 2.10 |
ENST00000335953.4 |
ZBTB16 |
zinc finger and BTB domain containing 16 |
| chr11_-_35547151 | 2.09 |
ENST00000378878.3 ENST00000529303.1 ENST00000278360.3 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr9_-_79520989 | 2.04 |
ENST00000376713.3 ENST00000376718.3 ENST00000428286.1 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr20_-_34042558 | 1.95 |
ENST00000374372.1 |
GDF5 |
growth differentiation factor 5 |
| chr5_+_156712372 | 1.86 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr4_+_145567297 | 1.83 |
ENST00000434550.2 |
HHIP |
hedgehog interacting protein |
| chr17_-_66951474 | 1.79 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr9_+_91606355 | 1.67 |
ENST00000358157.2 |
S1PR3 |
sphingosine-1-phosphate receptor 3 |
| chr22_+_38864041 | 1.67 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr4_-_39640513 | 1.46 |
ENST00000511809.1 ENST00000505729.1 |
SMIM14 |
small integral membrane protein 14 |
| chr22_-_20850128 | 1.46 |
ENST00000328879.4 |
KLHL22 |
kelch-like family member 22 |
| chr14_+_96505659 | 1.40 |
ENST00000555004.1 |
C14orf132 |
chromosome 14 open reading frame 132 |
| chr7_-_19157248 | 1.32 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chr4_-_39640700 | 1.29 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chr8_+_97657449 | 1.13 |
ENST00000220763.5 |
CPQ |
carboxypeptidase Q |
| chr12_+_117176090 | 1.11 |
ENST00000257575.4 ENST00000407967.3 ENST00000392549.2 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr1_+_221051699 | 1.05 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chrX_-_101397433 | 1.03 |
ENST00000372774.3 |
TCEAL6 |
transcription elongation factor A (SII)-like 6 |
| chr5_-_132299290 | 1.00 |
ENST00000378595.3 |
AFF4 |
AF4/FMR2 family, member 4 |
| chr9_+_36036430 | 0.96 |
ENST00000377966.3 |
RECK |
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr13_-_49107303 | 0.96 |
ENST00000344532.3 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr9_-_89562104 | 0.92 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chrX_+_102862834 | 0.90 |
ENST00000372627.5 ENST00000243286.3 |
TCEAL3 |
transcription elongation factor A (SII)-like 3 |
| chr19_+_49458107 | 0.89 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chr16_-_73082274 | 0.88 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr19_+_35630628 | 0.88 |
ENST00000588715.1 ENST00000588607.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr16_+_53164833 | 0.86 |
ENST00000564845.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr12_+_117176113 | 0.85 |
ENST00000319176.7 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr19_+_10527449 | 0.85 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr19_+_35630926 | 0.82 |
ENST00000588081.1 ENST00000589121.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr21_+_38445539 | 0.81 |
ENST00000418766.1 ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3 |
tetratricopeptide repeat domain 3 |
| chr12_-_76953453 | 0.81 |
ENST00000549570.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr7_+_102073966 | 0.77 |
ENST00000495936.1 ENST00000356387.2 ENST00000478730.2 ENST00000468241.1 ENST00000403646.3 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
| chr6_-_25042231 | 0.71 |
ENST00000510784.2 |
FAM65B |
family with sequence similarity 65, member B |
| chr22_-_31741757 | 0.70 |
ENST00000215919.3 |
PATZ1 |
POZ (BTB) and AT hook containing zinc finger 1 |
| chr14_-_23822080 | 0.67 |
ENST00000397267.1 ENST00000354772.3 |
SLC22A17 |
solute carrier family 22, member 17 |
| chr12_+_1100449 | 0.67 |
ENST00000360905.4 |
ERC1 |
ELKS/RAB6-interacting/CAST family member 1 |
| chr6_+_132891461 | 0.67 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr14_-_23822061 | 0.67 |
ENST00000397260.3 |
SLC22A17 |
solute carrier family 22, member 17 |
| chr7_-_98467629 | 0.66 |
ENST00000339375.4 |
TMEM130 |
transmembrane protein 130 |
| chrX_+_153672468 | 0.64 |
ENST00000393600.3 |
FAM50A |
family with sequence similarity 50, member A |
| chr7_-_98467543 | 0.64 |
ENST00000345589.4 |
TMEM130 |
transmembrane protein 130 |
| chr7_-_8302207 | 0.64 |
ENST00000407906.1 |
ICA1 |
islet cell autoantigen 1, 69kDa |
| chr7_-_98467489 | 0.62 |
ENST00000416379.2 |
TMEM130 |
transmembrane protein 130 |
| chr1_-_15850676 | 0.62 |
ENST00000440484.1 ENST00000333868.5 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr12_+_56661033 | 0.62 |
ENST00000433805.2 |
COQ10A |
coenzyme Q10 homolog A (S. cerevisiae) |
| chr20_+_34043085 | 0.62 |
ENST00000397527.1 ENST00000342580.4 |
CEP250 |
centrosomal protein 250kDa |
| chr3_+_129159039 | 0.60 |
ENST00000507564.1 ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr8_+_22422749 | 0.59 |
ENST00000523900.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr3_+_129158926 | 0.59 |
ENST00000347300.2 ENST00000296266.3 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr1_-_15850839 | 0.57 |
ENST00000348549.5 ENST00000546424.1 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr12_-_110011288 | 0.57 |
ENST00000540016.1 ENST00000266839.5 |
MMAB |
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr20_-_34638841 | 0.56 |
ENST00000565493.1 |
LINC00657 |
long intergenic non-protein coding RNA 657 |
| chrX_-_100872911 | 0.55 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr12_+_56660633 | 0.55 |
ENST00000308197.5 |
COQ10A |
coenzyme Q10 homolog A (S. cerevisiae) |
| chr5_+_126112794 | 0.53 |
ENST00000261366.5 ENST00000395354.1 |
LMNB1 |
lamin B1 |
| chr5_-_131562935 | 0.53 |
ENST00000379104.2 ENST00000379100.2 ENST00000428369.1 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr12_+_72148614 | 0.52 |
ENST00000261263.3 |
RAB21 |
RAB21, member RAS oncogene family |
| chr15_+_67420441 | 0.51 |
ENST00000558894.1 |
SMAD3 |
SMAD family member 3 |
| chr22_-_20850070 | 0.51 |
ENST00000440659.2 ENST00000458248.1 ENST00000443285.1 ENST00000444967.1 ENST00000451553.1 ENST00000431430.1 |
KLHL22 |
kelch-like family member 22 |
| chr15_+_41913690 | 0.50 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chr8_+_22423219 | 0.49 |
ENST00000523965.1 ENST00000521554.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr13_+_114238997 | 0.48 |
ENST00000538138.1 ENST00000375370.5 |
TFDP1 |
transcription factor Dp-1 |
| chr20_-_2821756 | 0.47 |
ENST00000356872.3 ENST00000439542.1 |
PCED1A |
PC-esterase domain containing 1A |
| chr20_-_2821271 | 0.46 |
ENST00000448755.1 ENST00000360652.2 |
PCED1A |
PC-esterase domain containing 1A |
| chr9_+_112542572 | 0.46 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr16_+_29973351 | 0.45 |
ENST00000602948.1 ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219 |
transmembrane protein 219 |
| chr10_-_118032979 | 0.44 |
ENST00000355422.6 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr22_-_36903101 | 0.44 |
ENST00000397224.4 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr7_-_100493744 | 0.44 |
ENST00000428317.1 ENST00000441605.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr11_-_77791156 | 0.43 |
ENST00000281031.4 |
NDUFC2 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr1_+_114472481 | 0.43 |
ENST00000369555.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chrX_+_48433326 | 0.43 |
ENST00000376755.1 |
RBM3 |
RNA binding motif (RNP1, RRM) protein 3 |
| chr6_-_24489842 | 0.43 |
ENST00000230036.1 |
GPLD1 |
glycosylphosphatidylinositol specific phospholipase D1 |
| chr19_-_18391708 | 0.42 |
ENST00000600972.1 |
JUND |
jun D proto-oncogene |
| chr12_-_56123444 | 0.42 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr1_-_208417620 | 0.42 |
ENST00000367033.3 |
PLXNA2 |
plexin A2 |
| chr14_-_35183755 | 0.41 |
ENST00000555765.1 |
CFL2 |
cofilin 2 (muscle) |
| chr17_+_66511540 | 0.41 |
ENST00000588188.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr4_-_169931231 | 0.41 |
ENST00000504561.1 |
CBR4 |
carbonyl reductase 4 |
| chr10_+_35416090 | 0.41 |
ENST00000354759.3 |
CREM |
cAMP responsive element modulator |
| chr14_-_35183886 | 0.40 |
ENST00000298159.6 |
CFL2 |
cofilin 2 (muscle) |
| chr20_+_39657454 | 0.40 |
ENST00000361337.2 |
TOP1 |
topoisomerase (DNA) I |
| chr20_+_56964169 | 0.40 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr16_-_54320675 | 0.40 |
ENST00000329734.3 |
IRX3 |
iroquois homeobox 3 |
| chr4_-_119757239 | 0.40 |
ENST00000280551.6 |
SEC24D |
SEC24 family member D |
| chr17_+_65821636 | 0.40 |
ENST00000544778.2 |
BPTF |
bromodomain PHD finger transcription factor |
| chr6_+_132873832 | 0.39 |
ENST00000275200.1 |
TAAR8 |
trace amine associated receptor 8 |
| chr15_+_96869165 | 0.39 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr15_-_40213080 | 0.39 |
ENST00000561100.1 |
GPR176 |
G protein-coupled receptor 176 |
| chr12_+_1100370 | 0.39 |
ENST00000543086.3 ENST00000546231.2 ENST00000397203.2 |
ERC1 |
ELKS/RAB6-interacting/CAST family member 1 |
| chr20_+_37590942 | 0.39 |
ENST00000373325.2 ENST00000252011.3 ENST00000373323.4 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr3_+_170075436 | 0.39 |
ENST00000476188.1 ENST00000259119.4 ENST00000426052.2 |
SKIL |
SKI-like oncogene |
| chr6_+_127588020 | 0.39 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr1_-_25573937 | 0.39 |
ENST00000417642.2 ENST00000431849.2 |
C1orf63 |
chromosome 1 open reading frame 63 |
| chr3_-_101395936 | 0.39 |
ENST00000461821.1 |
ZBTB11 |
zinc finger and BTB domain containing 11 |
| chr1_-_160313025 | 0.38 |
ENST00000368069.3 ENST00000241704.7 |
COPA |
coatomer protein complex, subunit alpha |
| chr12_-_76953513 | 0.38 |
ENST00000547540.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr5_-_64777733 | 0.38 |
ENST00000381055.3 |
ADAMTS6 |
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr19_+_16186903 | 0.38 |
ENST00000588507.1 |
TPM4 |
tropomyosin 4 |
| chr1_-_25573977 | 0.38 |
ENST00000243189.7 |
C1orf63 |
chromosome 1 open reading frame 63 |
| chrX_+_102631844 | 0.38 |
ENST00000372634.1 ENST00000299872.7 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr10_+_35415719 | 0.38 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr10_-_104953009 | 0.37 |
ENST00000470299.1 ENST00000343289.5 |
NT5C2 |
5'-nucleotidase, cytosolic II |
| chrX_+_102631248 | 0.37 |
ENST00000361298.4 ENST00000372645.3 ENST00000372635.1 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr4_+_668348 | 0.37 |
ENST00000511290.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr14_+_55518349 | 0.36 |
ENST00000395468.4 |
MAPK1IP1L |
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr5_+_133984462 | 0.35 |
ENST00000398844.2 ENST00000322887.4 |
SEC24A |
SEC24 family member A |
| chr5_-_171881491 | 0.35 |
ENST00000311601.5 |
SH3PXD2B |
SH3 and PX domains 2B |
| chr1_-_51425902 | 0.35 |
ENST00000396153.2 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr1_+_197170592 | 0.35 |
ENST00000535699.1 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr17_+_4046462 | 0.34 |
ENST00000577075.2 ENST00000575251.1 ENST00000301391.3 |
CYB5D2 |
cytochrome b5 domain containing 2 |
| chr7_+_100464760 | 0.34 |
ENST00000200457.4 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr3_+_3168692 | 0.34 |
ENST00000402675.1 ENST00000413000.1 |
TRNT1 |
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chrX_-_102941596 | 0.33 |
ENST00000441076.2 ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr2_+_242254507 | 0.33 |
ENST00000391973.2 |
SEPT2 |
septin 2 |
| chr3_+_44771088 | 0.33 |
ENST00000396048.2 |
ZNF501 |
zinc finger protein 501 |
| chr20_-_42815733 | 0.33 |
ENST00000342272.3 |
JPH2 |
junctophilin 2 |
| chr6_+_127587755 | 0.33 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr6_-_90062543 | 0.33 |
ENST00000435041.2 |
UBE2J1 |
ubiquitin-conjugating enzyme E2, J1 |
| chr6_+_33168637 | 0.33 |
ENST00000374677.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr12_-_102455846 | 0.32 |
ENST00000545679.1 |
CCDC53 |
coiled-coil domain containing 53 |
| chr12_-_102455902 | 0.32 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr15_-_43785274 | 0.32 |
ENST00000413546.1 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr6_+_33168597 | 0.31 |
ENST00000374675.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr7_+_99102267 | 0.31 |
ENST00000326775.5 ENST00000451158.1 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr12_-_10766184 | 0.31 |
ENST00000539554.1 ENST00000381881.2 ENST00000320756.2 |
MAGOHB |
mago-nashi homolog B (Drosophila) |
| chr17_-_57184260 | 0.30 |
ENST00000376149.3 ENST00000393066.3 |
TRIM37 |
tripartite motif containing 37 |
| chr2_-_27712583 | 0.30 |
ENST00000260570.3 ENST00000359466.6 ENST00000416524.2 |
IFT172 |
intraflagellar transport 172 homolog (Chlamydomonas) |
| chrX_+_108780347 | 0.30 |
ENST00000372103.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr12_+_113796347 | 0.30 |
ENST00000545182.2 ENST00000280800.3 |
PLBD2 |
phospholipase B domain containing 2 |
| chr8_-_38126635 | 0.30 |
ENST00000529359.1 |
PPAPDC1B |
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr6_+_29624898 | 0.30 |
ENST00000396704.3 ENST00000483013.1 ENST00000490427.1 ENST00000416766.2 ENST00000376891.4 ENST00000376898.3 ENST00000396701.2 ENST00000494692.1 ENST00000431798.2 |
MOG |
myelin oligodendrocyte glycoprotein |
| chr5_-_131563501 | 0.29 |
ENST00000401867.1 ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr15_-_50978965 | 0.29 |
ENST00000560955.1 ENST00000313478.7 |
TRPM7 |
transient receptor potential cation channel, subfamily M, member 7 |
| chr15_+_76135622 | 0.29 |
ENST00000338677.4 ENST00000267938.4 ENST00000569423.1 |
UBE2Q2 |
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr6_-_84937314 | 0.29 |
ENST00000257766.4 ENST00000403245.3 |
KIAA1009 |
KIAA1009 |
| chr8_-_38126675 | 0.29 |
ENST00000531823.1 ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B |
phosphatidic acid phosphatase type 2 domain containing 1B |
| chrX_+_134478706 | 0.29 |
ENST00000370761.3 ENST00000339249.4 ENST00000370760.3 |
ZNF449 |
zinc finger protein 449 |
| chr2_-_242212227 | 0.29 |
ENST00000427007.1 ENST00000458564.1 ENST00000452065.1 ENST00000427183.2 ENST00000426343.1 ENST00000422080.1 ENST00000449504.1 ENST00000449864.1 ENST00000391975.1 |
HDLBP |
high density lipoprotein binding protein |
| chr7_-_27169801 | 0.28 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr9_+_100000717 | 0.28 |
ENST00000375205.2 ENST00000357054.1 ENST00000395220.1 ENST00000375202.2 ENST00000411667.2 |
CCDC180 |
coiled-coil domain containing 180 |
| chr6_+_29624758 | 0.28 |
ENST00000376917.3 ENST00000376902.3 ENST00000533330.2 ENST00000376888.2 |
MOG |
myelin oligodendrocyte glycoprotein |
| chr19_+_5904866 | 0.27 |
ENST00000339485.3 |
VMAC |
vimentin-type intermediate filament associated coiled-coil protein |
| chr10_-_31320840 | 0.27 |
ENST00000375311.1 |
ZNF438 |
zinc finger protein 438 |
| chr2_-_10588630 | 0.27 |
ENST00000234111.4 |
ODC1 |
ornithine decarboxylase 1 |
| chr20_+_44421137 | 0.27 |
ENST00000415790.1 |
DNTTIP1 |
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr10_+_35415978 | 0.27 |
ENST00000429130.3 ENST00000469949.2 ENST00000460270.1 |
CREM |
cAMP responsive element modulator |
| chr12_-_76953573 | 0.27 |
ENST00000549646.1 ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr12_+_54718904 | 0.27 |
ENST00000262061.2 ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
| chr1_+_32645269 | 0.26 |
ENST00000373610.3 |
TXLNA |
taxilin alpha |
| chr11_+_120195992 | 0.26 |
ENST00000314475.2 ENST00000529187.1 |
TMEM136 |
transmembrane protein 136 |
| chr14_+_50087468 | 0.26 |
ENST00000305386.2 |
MGAT2 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr9_+_99212403 | 0.26 |
ENST00000375251.3 ENST00000375249.4 |
HABP4 |
hyaluronan binding protein 4 |
| chr20_+_2821340 | 0.26 |
ENST00000380445.3 ENST00000380469.3 |
VPS16 |
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr1_-_145039949 | 0.26 |
ENST00000313382.9 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr11_+_1891380 | 0.26 |
ENST00000429923.1 ENST00000418975.1 ENST00000406638.2 |
LSP1 |
lymphocyte-specific protein 1 |
| chr9_+_126777676 | 0.26 |
ENST00000488674.2 |
LHX2 |
LIM homeobox 2 |
| chr7_+_99102573 | 0.25 |
ENST00000394170.2 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr7_+_97910981 | 0.25 |
ENST00000297290.3 |
BRI3 |
brain protein I3 |
| chr5_-_114961673 | 0.25 |
ENST00000333314.3 |
TMED7-TICAM2 |
TMED7-TICAM2 readthrough |
| chr19_+_13858593 | 0.25 |
ENST00000221554.8 |
CCDC130 |
coiled-coil domain containing 130 |
| chr11_+_58910295 | 0.25 |
ENST00000420244.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr17_+_57642886 | 0.25 |
ENST00000251241.4 ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr12_-_125398602 | 0.25 |
ENST00000541272.1 ENST00000535131.1 |
UBC |
ubiquitin C |
| chr2_-_211036051 | 0.25 |
ENST00000418791.1 ENST00000452086.1 ENST00000281772.9 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
| chr10_-_22292675 | 0.24 |
ENST00000376946.1 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr19_-_41196534 | 0.24 |
ENST00000252891.4 |
NUMBL |
numb homolog (Drosophila)-like |
| chr20_+_44420617 | 0.24 |
ENST00000449078.1 ENST00000456939.1 |
DNTTIP1 |
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr5_-_132299313 | 0.24 |
ENST00000265343.5 |
AFF4 |
AF4/FMR2 family, member 4 |
| chr17_+_37026284 | 0.24 |
ENST00000433206.2 ENST00000435347.3 |
LASP1 |
LIM and SH3 protein 1 |
| chr14_-_77495007 | 0.24 |
ENST00000238647.3 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
| chr10_+_35416223 | 0.24 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr1_+_26798955 | 0.24 |
ENST00000361427.5 |
HMGN2 |
high mobility group nucleosomal binding domain 2 |
| chr13_-_36920872 | 0.24 |
ENST00000451493.1 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr17_+_33914460 | 0.24 |
ENST00000537622.2 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr5_+_60628074 | 0.23 |
ENST00000252744.5 |
ZSWIM6 |
zinc finger, SWIM-type containing 6 |
| chr20_+_35090150 | 0.23 |
ENST00000340491.4 |
DLGAP4 |
discs, large (Drosophila) homolog-associated protein 4 |
| chr19_-_48018203 | 0.23 |
ENST00000595227.1 ENST00000593761.1 ENST00000263354.3 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr6_+_29624862 | 0.23 |
ENST00000376894.4 |
MOG |
myelin oligodendrocyte glycoprotein |
| chr3_+_50284321 | 0.23 |
ENST00000451956.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr7_+_99699280 | 0.23 |
ENST00000421755.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr5_-_56247935 | 0.22 |
ENST00000381199.3 ENST00000381226.3 ENST00000381213.3 |
MIER3 |
mesoderm induction early response 1, family member 3 |
| chr11_-_77790865 | 0.22 |
ENST00000534029.1 ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2 NDUFC2-KCTD14 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr5_-_94890648 | 0.22 |
ENST00000513823.1 ENST00000514952.1 ENST00000358746.2 |
TTC37 |
tetratricopeptide repeat domain 37 |
| chr9_-_124855885 | 0.22 |
ENST00000321582.5 ENST00000373776.3 |
TTLL11 |
tubulin tyrosine ligase-like family, member 11 |
| chr9_-_127703333 | 0.22 |
ENST00000373555.4 |
GOLGA1 |
golgin A1 |
| chrX_+_108780062 | 0.22 |
ENST00000372106.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr7_+_97910962 | 0.22 |
ENST00000539286.1 |
BRI3 |
brain protein I3 |
| chr8_+_22423168 | 0.22 |
ENST00000518912.1 ENST00000428103.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr3_+_184529929 | 0.21 |
ENST00000287546.4 ENST00000437079.3 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr3_+_184053703 | 0.21 |
ENST00000450976.1 ENST00000418281.1 ENST00000340957.5 ENST00000433578.1 |
FAM131A |
family with sequence similarity 131, member A |
| chr19_-_46088068 | 0.21 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr20_+_44420570 | 0.21 |
ENST00000372622.3 |
DNTTIP1 |
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr17_+_59529743 | 0.21 |
ENST00000589003.1 ENST00000393853.4 |
TBX4 |
T-box 4 |
| chr9_+_131038425 | 0.21 |
ENST00000320188.5 ENST00000608796.1 ENST00000419867.2 ENST00000418976.1 |
SWI5 |
SWI5 recombination repair homolog (yeast) |
| chr3_+_184529948 | 0.21 |
ENST00000436792.2 ENST00000446204.2 ENST00000422105.1 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.4 | 2.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.4 | 2.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 0.9 | GO:1903519 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.3 | 1.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.3 | 1.7 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.3 | 2.0 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.3 | 1.3 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 0.9 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 1.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 1.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 0.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 0.6 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 1.8 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.6 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.4 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 1.0 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 1.5 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 1.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.4 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 4.1 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.1 | 0.4 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.3 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.3 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 1.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.2 | GO:0003193 | pulmonary valve formation(GO:0003193) visceral motor neuron differentiation(GO:0021524) foramen ovale closure(GO:0035922) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.2 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 1.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 1.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.2 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.2 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.5 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 2.0 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.6 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.3 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.3 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.3 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.4 | GO:0044598 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.4 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.4 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.8 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.4 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.1 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) negative regulation by host of symbiont molecular function(GO:0052405) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.0 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.2 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.7 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 1.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.0 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:1903984 | negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.0 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.2 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0043928 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.0 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 1.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.2 | 2.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 1.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 1.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.5 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.2 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 1.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 4.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0044423 | virion(GO:0019012) virion part(GO:0044423) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.7 | GO:0030133 | transport vesicle(GO:0030133) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 1.8 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 1.8 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 1.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 1.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 2.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.4 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 1.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 1.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.2 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 1.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0030552 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 5.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 3.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |