Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
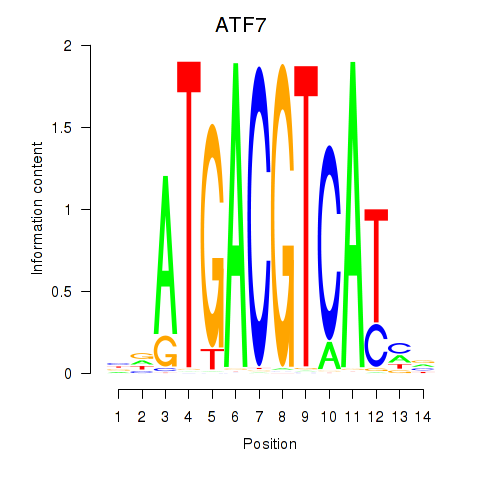
Results for ATF7
Z-value: 0.47
Transcription factors associated with ATF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF7
|
ENSG00000170653.14 | ATF7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF7 | hg19_v2_chr12_-_54019755_54019804 | -0.69 | 6.0e-02 | Click! |
Activity profile of ATF7 motif
Sorted Z-values of ATF7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_156030937 | 0.90 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chr14_+_68086515 | 0.85 |
ENST00000261783.3 |
ARG2 |
arginase 2 |
| chr6_+_28048753 | 0.58 |
ENST00000377325.1 |
ZNF165 |
zinc finger protein 165 |
| chr14_+_96671016 | 0.53 |
ENST00000542454.2 ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2 RP11-404P21.8 |
bradykinin receptor B2 Uncharacterized protein |
| chr4_+_75310851 | 0.50 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr4_+_75311019 | 0.50 |
ENST00000502307.1 |
AREG |
amphiregulin |
| chr4_+_85504075 | 0.48 |
ENST00000295887.5 |
CDS1 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr11_-_118122996 | 0.41 |
ENST00000525386.1 ENST00000527472.1 ENST00000278949.4 |
MPZL3 |
myelin protein zero-like 3 |
| chr12_-_12715266 | 0.41 |
ENST00000228862.2 |
DUSP16 |
dual specificity phosphatase 16 |
| chr1_+_2004901 | 0.39 |
ENST00000400921.2 |
PRKCZ |
protein kinase C, zeta |
| chr1_+_2005425 | 0.39 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr11_-_77185094 | 0.37 |
ENST00000278568.4 ENST00000356341.3 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr12_-_52604607 | 0.35 |
ENST00000551894.1 ENST00000553017.1 |
C12orf80 |
chromosome 12 open reading frame 80 |
| chr4_+_75480629 | 0.34 |
ENST00000380846.3 |
AREGB |
amphiregulin B |
| chr20_-_1306391 | 0.34 |
ENST00000339987.3 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chr20_-_1306351 | 0.33 |
ENST00000381812.1 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chr11_-_62389449 | 0.30 |
ENST00000534026.1 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr12_-_31882108 | 0.29 |
ENST00000281471.6 |
AMN1 |
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr14_+_96722539 | 0.28 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr5_-_175964366 | 0.27 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr7_-_142247606 | 0.26 |
ENST00000390361.3 |
TRBV7-3 |
T cell receptor beta variable 7-3 |
| chr18_+_23806437 | 0.26 |
ENST00000578121.1 |
TAF4B |
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr7_-_140624499 | 0.24 |
ENST00000288602.6 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr19_-_56826157 | 0.23 |
ENST00000592509.1 ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
| chr5_-_95297534 | 0.23 |
ENST00000513343.1 ENST00000431061.2 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr11_-_123525289 | 0.23 |
ENST00000392770.2 ENST00000299333.3 ENST00000530277.1 |
SCN3B |
sodium channel, voltage-gated, type III, beta subunit |
| chr22_+_25003568 | 0.22 |
ENST00000447416.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chrX_+_48398053 | 0.22 |
ENST00000537536.1 ENST00000418627.1 |
TBC1D25 |
TBC1 domain family, member 25 |
| chr17_-_39780819 | 0.21 |
ENST00000311208.8 |
KRT17 |
keratin 17 |
| chr8_-_95274536 | 0.21 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr18_-_10787140 | 0.21 |
ENST00000383408.2 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr8_+_98788003 | 0.20 |
ENST00000521545.2 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr13_-_101327028 | 0.20 |
ENST00000328767.5 ENST00000342624.5 ENST00000376234.3 ENST00000423847.1 |
TMTC4 |
transmembrane and tetratricopeptide repeat containing 4 |
| chr19_+_45504688 | 0.19 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr14_+_96722152 | 0.19 |
ENST00000216629.6 |
BDKRB1 |
bradykinin receptor B1 |
| chr8_+_98788057 | 0.19 |
ENST00000517924.1 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr1_+_26496362 | 0.19 |
ENST00000374266.5 ENST00000270812.5 |
ZNF593 |
zinc finger protein 593 |
| chr16_+_68057153 | 0.18 |
ENST00000358896.6 ENST00000568099.2 |
DUS2 |
dihydrouridine synthase 2 |
| chr12_-_64616019 | 0.18 |
ENST00000311915.8 ENST00000398055.3 ENST00000544871.1 |
C12orf66 |
chromosome 12 open reading frame 66 |
| chr17_-_7155802 | 0.18 |
ENST00000572043.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr8_-_10697281 | 0.18 |
ENST00000524114.1 ENST00000553390.1 ENST00000554914.1 |
PINX1 SOX7 SOX7 |
PIN2/TERF1 interacting, telomerase inhibitor 1 SRY (sex determining region Y)-box 7 Transcription factor SOX-7; Uncharacterized protein; cDNA FLJ58508, highly similar to Transcription factor SOX-7 |
| chr17_-_7155274 | 0.18 |
ENST00000318988.6 ENST00000575783.1 ENST00000573600.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr16_+_68056844 | 0.17 |
ENST00000565263.1 |
DUS2 |
dihydrouridine synthase 2 |
| chr16_+_68057179 | 0.17 |
ENST00000567100.1 ENST00000432752.1 ENST00000569289.1 ENST00000564781.1 |
DUS2 |
dihydrouridine synthase 2 |
| chr17_-_8151353 | 0.17 |
ENST00000315684.8 |
CTC1 |
CTS telomere maintenance complex component 1 |
| chr4_+_128802016 | 0.16 |
ENST00000270861.5 ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4 |
polo-like kinase 4 |
| chr2_+_70142189 | 0.16 |
ENST00000264444.2 |
MXD1 |
MAX dimerization protein 1 |
| chr7_-_142120321 | 0.16 |
ENST00000390377.1 |
TRBV7-7 |
T cell receptor beta variable 7-7 |
| chr6_+_37225540 | 0.16 |
ENST00000373491.3 |
TBC1D22B |
TBC1 domain family, member 22B |
| chr9_-_77643189 | 0.15 |
ENST00000376837.3 |
C9orf41 |
chromosome 9 open reading frame 41 |
| chr17_-_7165662 | 0.15 |
ENST00000571881.2 ENST00000360325.7 |
CLDN7 |
claudin 7 |
| chr1_+_153750622 | 0.15 |
ENST00000532853.1 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr12_+_112856690 | 0.14 |
ENST00000392597.1 ENST00000351677.2 |
PTPN11 |
protein tyrosine phosphatase, non-receptor type 11 |
| chr3_+_158288942 | 0.14 |
ENST00000491767.1 ENST00000355893.5 |
MLF1 |
myeloid leukemia factor 1 |
| chr7_+_30174426 | 0.14 |
ENST00000324453.8 |
C7orf41 |
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr18_+_76829441 | 0.14 |
ENST00000458297.2 |
ATP9B |
ATPase, class II, type 9B |
| chr9_+_131644781 | 0.14 |
ENST00000259324.5 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr9_+_131644388 | 0.13 |
ENST00000372600.4 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr6_-_19804973 | 0.13 |
ENST00000457670.1 ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2 |
RP4-625H18.2 |
| chr5_-_16509101 | 0.13 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr19_-_50979981 | 0.13 |
ENST00000595790.1 ENST00000600100.1 |
FAM71E1 |
family with sequence similarity 71, member E1 |
| chr9_-_131644202 | 0.13 |
ENST00000320665.6 ENST00000436267.2 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr9_-_131644306 | 0.13 |
ENST00000302586.3 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr16_-_4588469 | 0.13 |
ENST00000588381.1 ENST00000563332.2 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr1_+_29213584 | 0.13 |
ENST00000343067.4 ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr4_+_1873100 | 0.13 |
ENST00000508803.1 |
WHSC1 |
Wolf-Hirschhorn syndrome candidate 1 |
| chr7_+_5632436 | 0.13 |
ENST00000340250.6 ENST00000382361.3 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chr9_+_131644398 | 0.13 |
ENST00000372599.3 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr13_-_31736478 | 0.12 |
ENST00000445273.2 |
HSPH1 |
heat shock 105kDa/110kDa protein 1 |
| chr19_+_10362882 | 0.12 |
ENST00000393733.2 ENST00000588502.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr16_-_3767551 | 0.12 |
ENST00000246957.5 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr7_+_142031986 | 0.12 |
ENST00000547918.2 |
TRBV7-1 |
T cell receptor beta variable 7-1 (non-functional) |
| chr5_-_95297678 | 0.12 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr11_-_85376121 | 0.12 |
ENST00000527447.1 |
CREBZF |
CREB/ATF bZIP transcription factor |
| chr16_-_3767506 | 0.12 |
ENST00000538171.1 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr9_-_77643307 | 0.12 |
ENST00000376834.3 ENST00000376830.3 |
C9orf41 |
chromosome 9 open reading frame 41 |
| chr22_+_25003606 | 0.12 |
ENST00000432867.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr16_-_4588822 | 0.12 |
ENST00000564828.1 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr21_+_43442100 | 0.11 |
ENST00000455701.1 ENST00000596595.1 |
ZNF295-AS1 |
ZNF295 antisense RNA 1 |
| chr19_+_13056663 | 0.11 |
ENST00000541222.1 ENST00000316856.3 ENST00000586534.1 ENST00000592268.1 |
RAD23A |
RAD23 homolog A (S. cerevisiae) |
| chr12_+_7014126 | 0.11 |
ENST00000415834.1 ENST00000436789.1 |
LRRC23 |
leucine rich repeat containing 23 |
| chr6_+_139456226 | 0.11 |
ENST00000367658.2 |
HECA |
headcase homolog (Drosophila) |
| chr4_+_159131346 | 0.11 |
ENST00000508243.1 ENST00000296529.6 |
TMEM144 |
transmembrane protein 144 |
| chr22_-_44258360 | 0.11 |
ENST00000330884.4 ENST00000249130.5 |
SULT4A1 |
sulfotransferase family 4A, member 1 |
| chr6_+_30029008 | 0.11 |
ENST00000332435.5 ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1 |
zinc ribbon domain containing 1 |
| chr17_-_43209862 | 0.11 |
ENST00000322765.5 |
PLCD3 |
phospholipase C, delta 3 |
| chr5_-_138739739 | 0.11 |
ENST00000514983.1 ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24 |
spermatogenesis associated 24 |
| chr3_+_158288960 | 0.11 |
ENST00000484955.1 ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1 |
myeloid leukemia factor 1 |
| chr17_+_41476327 | 0.11 |
ENST00000320033.4 |
ARL4D |
ADP-ribosylation factor-like 4D |
| chr6_-_91006461 | 0.10 |
ENST00000257749.4 ENST00000343122.3 ENST00000406998.2 ENST00000453877.1 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr19_+_18723660 | 0.10 |
ENST00000262817.3 |
TMEM59L |
transmembrane protein 59-like |
| chr1_+_29213678 | 0.10 |
ENST00000347529.3 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr9_-_2844058 | 0.10 |
ENST00000397885.2 |
KIAA0020 |
KIAA0020 |
| chr16_+_19078960 | 0.10 |
ENST00000568985.1 ENST00000566110.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr2_-_220264703 | 0.10 |
ENST00000519905.1 ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP |
aspartyl aminopeptidase |
| chr5_-_131892501 | 0.09 |
ENST00000450655.1 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr19_+_8455077 | 0.09 |
ENST00000328024.6 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr16_-_86588627 | 0.09 |
ENST00000565482.1 ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD |
methenyltetrahydrofolate synthetase domain containing |
| chr11_-_62389621 | 0.09 |
ENST00000531383.1 ENST00000265471.5 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr4_-_122744998 | 0.09 |
ENST00000274026.5 |
CCNA2 |
cyclin A2 |
| chr16_+_19079215 | 0.09 |
ENST00000544894.2 ENST00000561858.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr9_+_125027127 | 0.09 |
ENST00000441707.1 ENST00000373723.5 ENST00000373729.1 |
MRRF |
mitochondrial ribosome recycling factor |
| chr19_+_59055814 | 0.09 |
ENST00000594806.1 ENST00000253024.5 ENST00000341753.6 |
TRIM28 |
tripartite motif containing 28 |
| chr11_+_28129795 | 0.09 |
ENST00000406787.3 ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15 |
methyltransferase like 15 |
| chr19_+_8455200 | 0.09 |
ENST00000601897.1 ENST00000594216.1 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr20_+_42295745 | 0.09 |
ENST00000396863.4 ENST00000217026.4 |
MYBL2 |
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr7_-_121944491 | 0.09 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr19_+_52693259 | 0.09 |
ENST00000322088.6 ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A |
protein phosphatase 2, regulatory subunit A, alpha |
| chr9_-_99382065 | 0.09 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chr1_-_16482554 | 0.09 |
ENST00000358432.5 |
EPHA2 |
EPH receptor A2 |
| chr4_-_186456766 | 0.09 |
ENST00000284771.6 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr19_-_49622348 | 0.08 |
ENST00000408991.2 |
C19orf73 |
chromosome 19 open reading frame 73 |
| chr4_-_113558014 | 0.08 |
ENST00000503172.1 ENST00000505019.1 ENST00000309071.5 |
C4orf21 |
chromosome 4 open reading frame 21 |
| chr14_-_102552659 | 0.08 |
ENST00000441629.2 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr4_-_2010562 | 0.08 |
ENST00000411649.1 ENST00000542778.1 ENST00000411638.2 ENST00000431323.1 |
NELFA |
negative elongation factor complex member A |
| chr4_-_186456652 | 0.08 |
ENST00000284767.5 ENST00000284770.5 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr16_-_4588762 | 0.08 |
ENST00000562334.1 ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr6_-_31774714 | 0.08 |
ENST00000375661.5 |
LSM2 |
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr6_+_26204825 | 0.08 |
ENST00000360441.4 |
HIST1H4E |
histone cluster 1, H4e |
| chr8_-_21966893 | 0.08 |
ENST00000522405.1 ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr15_-_72612470 | 0.08 |
ENST00000287202.5 |
CELF6 |
CUGBP, Elav-like family member 6 |
| chr16_+_19079311 | 0.08 |
ENST00000569127.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr7_-_31380502 | 0.08 |
ENST00000297142.3 |
NEUROD6 |
neuronal differentiation 6 |
| chrX_+_77359671 | 0.07 |
ENST00000373316.4 |
PGK1 |
phosphoglycerate kinase 1 |
| chr19_+_50180317 | 0.07 |
ENST00000534465.1 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr2_-_220408430 | 0.07 |
ENST00000243776.6 |
CHPF |
chondroitin polymerizing factor |
| chr10_+_123923205 | 0.07 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chrX_+_77359726 | 0.07 |
ENST00000442431.1 |
PGK1 |
phosphoglycerate kinase 1 |
| chr7_-_129592471 | 0.07 |
ENST00000473814.2 ENST00000490974.1 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
| chr10_+_123923105 | 0.07 |
ENST00000368999.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr10_+_123922941 | 0.07 |
ENST00000360561.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr19_-_55574538 | 0.07 |
ENST00000415061.3 |
RDH13 |
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr11_+_108535752 | 0.07 |
ENST00000322536.3 |
DDX10 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr8_-_70016408 | 0.07 |
ENST00000518540.1 |
RP11-600K15.1 |
RP11-600K15.1 |
| chr6_-_91006627 | 0.07 |
ENST00000537989.1 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr17_-_37009882 | 0.07 |
ENST00000378096.3 ENST00000394332.1 ENST00000394333.1 ENST00000577407.1 ENST00000479035.2 |
RPL23 |
ribosomal protein L23 |
| chr9_-_130477912 | 0.07 |
ENST00000543175.1 |
PTRH1 |
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr19_+_10362577 | 0.07 |
ENST00000592514.1 ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr17_+_7835419 | 0.07 |
ENST00000576538.1 ENST00000380262.3 ENST00000563694.1 ENST00000380255.3 ENST00000570782.1 |
CNTROB |
centrobin, centrosomal BRCA2 interacting protein |
| chr1_-_713985 | 0.07 |
ENST00000428504.1 |
RP11-206L10.2 |
RP11-206L10.2 |
| chr9_+_125026882 | 0.07 |
ENST00000297908.3 ENST00000373730.3 ENST00000546115.1 ENST00000344641.3 |
MRRF |
mitochondrial ribosome recycling factor |
| chr10_+_134351319 | 0.07 |
ENST00000368594.3 ENST00000368593.3 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
| chr22_+_32340447 | 0.06 |
ENST00000248975.5 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr3_-_51909600 | 0.06 |
ENST00000446461.1 |
IQCF5 |
IQ motif containing F5 |
| chr4_-_146019287 | 0.06 |
ENST00000502847.1 ENST00000513054.1 |
ANAPC10 |
anaphase promoting complex subunit 10 |
| chr2_+_3705785 | 0.06 |
ENST00000252505.3 |
ALLC |
allantoicase |
| chr19_+_36036583 | 0.06 |
ENST00000392205.1 |
TMEM147 |
transmembrane protein 147 |
| chr22_+_38597889 | 0.06 |
ENST00000338483.2 ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr3_-_33759699 | 0.06 |
ENST00000399362.4 ENST00000359576.5 ENST00000307312.7 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr6_-_32557610 | 0.06 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr3_+_10068095 | 0.06 |
ENST00000287647.3 ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2 |
Fanconi anemia, complementation group D2 |
| chr8_+_26149007 | 0.06 |
ENST00000380737.3 ENST00000524169.1 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr17_+_7155556 | 0.06 |
ENST00000570500.1 ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr12_-_100660833 | 0.06 |
ENST00000551642.1 ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4 |
DEP domain containing 4 |
| chr8_-_10697370 | 0.06 |
ENST00000314787.3 ENST00000426190.2 ENST00000519088.1 |
PINX1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr17_+_68071389 | 0.06 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_-_33759541 | 0.06 |
ENST00000468888.2 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr17_+_7155343 | 0.06 |
ENST00000573513.1 ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr16_+_11343475 | 0.06 |
ENST00000572173.1 |
RMI2 |
RecQ mediated genome instability 2 |
| chr3_+_51851612 | 0.06 |
ENST00000456080.1 |
IQCF3 |
IQ motif containing F3 |
| chr19_-_49122384 | 0.06 |
ENST00000550973.1 ENST00000550645.1 ENST00000549273.1 ENST00000552588.1 |
RPL18 |
ribosomal protein L18 |
| chr4_-_146019693 | 0.06 |
ENST00000514390.1 |
ANAPC10 |
anaphase promoting complex subunit 10 |
| chr16_+_2563871 | 0.06 |
ENST00000330398.4 ENST00000568562.1 ENST00000569317.1 |
ATP6V0C ATP6C |
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c Uncharacterized protein |
| chr7_-_124569991 | 0.06 |
ENST00000446993.1 ENST00000357628.3 ENST00000393329.1 |
POT1 |
protection of telomeres 1 |
| chr5_+_99871004 | 0.06 |
ENST00000312637.4 |
FAM174A |
family with sequence similarity 174, member A |
| chr6_+_150690028 | 0.06 |
ENST00000229447.5 ENST00000344419.3 |
IYD |
iodotyrosine deiodinase |
| chr17_+_29421987 | 0.06 |
ENST00000431387.4 |
NF1 |
neurofibromin 1 |
| chr19_+_36120009 | 0.06 |
ENST00000589871.1 |
RBM42 |
RNA binding motif protein 42 |
| chr1_-_32110467 | 0.06 |
ENST00000440872.2 ENST00000373703.4 |
PEF1 |
penta-EF-hand domain containing 1 |
| chr1_+_6685228 | 0.06 |
ENST00000307896.6 ENST00000377627.3 ENST00000472925.1 |
THAP3 |
THAP domain containing, apoptosis associated protein 3 |
| chr7_+_56019486 | 0.05 |
ENST00000446692.1 ENST00000285298.4 ENST00000443449.1 |
GBAS MRPS17 |
glioblastoma amplified sequence mitochondrial ribosomal protein S17 |
| chr17_+_29421900 | 0.05 |
ENST00000358273.4 ENST00000356175.3 |
NF1 |
neurofibromin 1 |
| chr16_-_3068171 | 0.05 |
ENST00000572154.1 ENST00000328796.4 |
CLDN6 |
claudin 6 |
| chr14_+_60715928 | 0.05 |
ENST00000395076.4 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr12_+_10365404 | 0.05 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr7_-_73153178 | 0.05 |
ENST00000437775.2 ENST00000222800.3 |
ABHD11 |
abhydrolase domain containing 11 |
| chr16_-_81040719 | 0.05 |
ENST00000219400.3 |
CMC2 |
C-x(9)-C motif containing 2 |
| chr2_-_112237835 | 0.05 |
ENST00000442293.1 ENST00000439494.1 |
MIR4435-1HG |
MIR4435-1 host gene (non-protein coding) |
| chr14_-_44976474 | 0.05 |
ENST00000340446.4 |
FSCB |
fibrous sheath CABYR binding protein |
| chr7_-_142176790 | 0.05 |
ENST00000390369.2 |
TRBV7-4 |
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr14_+_100705322 | 0.05 |
ENST00000262238.4 |
YY1 |
YY1 transcription factor |
| chr7_+_129984630 | 0.05 |
ENST00000355388.3 ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5 |
carboxypeptidase A5 |
| chr22_+_25003626 | 0.05 |
ENST00000451366.1 ENST00000406383.2 ENST00000428855.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr8_+_94929168 | 0.05 |
ENST00000518107.1 ENST00000396200.3 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr3_+_158288999 | 0.05 |
ENST00000482628.1 ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1 |
myeloid leukemia factor 1 |
| chr16_-_27561209 | 0.04 |
ENST00000356183.4 ENST00000561623.1 |
GTF3C1 |
general transcription factor IIIC, polypeptide 1, alpha 220kDa |
| chr17_+_6918354 | 0.04 |
ENST00000552775.1 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr17_+_68071458 | 0.04 |
ENST00000589377.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_+_6918064 | 0.04 |
ENST00000546760.1 ENST00000552402.1 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chrX_+_49832231 | 0.04 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr11_-_28129656 | 0.04 |
ENST00000263181.6 |
KIF18A |
kinesin family member 18A |
| chr17_+_6918093 | 0.04 |
ENST00000439424.2 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr12_-_93836028 | 0.04 |
ENST00000318066.2 |
UBE2N |
ubiquitin-conjugating enzyme E2N |
| chr3_-_52312636 | 0.04 |
ENST00000296490.3 |
WDR82 |
WD repeat domain 82 |
| chr4_-_48136217 | 0.04 |
ENST00000264316.4 |
TXK |
TXK tyrosine kinase |
| chr22_+_32340481 | 0.04 |
ENST00000397492.1 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr12_+_7014064 | 0.04 |
ENST00000443597.2 |
LRRC23 |
leucine rich repeat containing 23 |
| chr19_+_36119975 | 0.04 |
ENST00000589559.1 ENST00000360475.4 |
RBM42 |
RNA binding motif protein 42 |
| chr2_-_190627481 | 0.04 |
ENST00000264151.5 ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1 |
O-sialoglycoprotein endopeptidase-like 1 |
| chr20_-_17949100 | 0.04 |
ENST00000431277.1 |
SNX5 |
sorting nexin 5 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.8 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.5 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.2 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.5 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.9 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.2 | GO:1904751 | positive regulation of telomeric DNA binding(GO:1904744) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.9 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.4 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.1 | GO:1901491 | axial mesoderm formation(GO:0048320) negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:1903026 | ribosomal large subunit export from nucleus(GO:0000055) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.2 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0043605 | allantoin metabolic process(GO:0000255) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.2 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.9 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.0 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |