Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for BARHL2
Z-value: 0.78
Transcription factors associated with BARHL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARHL2
|
ENSG00000143032.7 | BARHL2 |
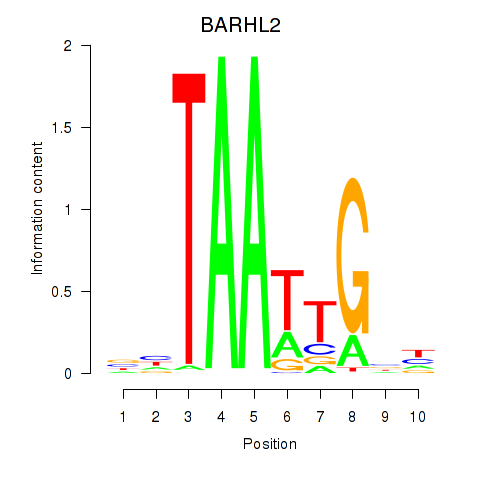
Activity profile of BARHL2 motif
Sorted Z-values of BARHL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BARHL2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91546926 | 1.88 |
ENST00000550758.1 |
DCN |
decorin |
| chr2_-_238322800 | 1.75 |
ENST00000392004.3 ENST00000433762.1 ENST00000347401.3 ENST00000353578.4 ENST00000346358.4 ENST00000392003.2 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr12_-_91573132 | 1.67 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_-_91576561 | 1.66 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr14_-_60097524 | 1.60 |
ENST00000342503.4 |
RTN1 |
reticulon 1 |
| chr14_-_60097297 | 1.52 |
ENST00000395090.1 |
RTN1 |
reticulon 1 |
| chr2_-_238323007 | 1.51 |
ENST00000295550.4 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr2_-_238322770 | 1.38 |
ENST00000472056.1 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr12_-_91576429 | 1.32 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr13_-_38172863 | 1.01 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr2_-_188419078 | 0.96 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_-_145277569 | 0.81 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr5_-_146781153 | 0.80 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr4_+_70916119 | 0.72 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chr1_-_232651312 | 0.68 |
ENST00000262861.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr5_+_95066823 | 0.65 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr10_-_49813090 | 0.63 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chrX_+_86772787 | 0.60 |
ENST00000373114.4 |
KLHL4 |
kelch-like family member 4 |
| chr5_-_20575959 | 0.59 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chrX_+_86772707 | 0.56 |
ENST00000373119.4 |
KLHL4 |
kelch-like family member 4 |
| chr2_+_152214098 | 0.55 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr13_-_67802549 | 0.54 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr5_-_111312622 | 0.51 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr2_+_33359646 | 0.50 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr10_+_69865866 | 0.50 |
ENST00000354393.2 |
MYPN |
myopalladin |
| chr17_+_59489112 | 0.50 |
ENST00000335108.2 |
C17orf82 |
chromosome 17 open reading frame 82 |
| chr15_-_37393406 | 0.44 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr9_+_99690592 | 0.42 |
ENST00000354649.3 |
NUTM2G |
NUT family member 2G |
| chr6_-_46293378 | 0.42 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr9_-_95244781 | 0.41 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr8_-_23712312 | 0.41 |
ENST00000290271.2 |
STC1 |
stanniocalcin 1 |
| chr11_+_6411670 | 0.41 |
ENST00000530395.1 ENST00000527275.1 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr11_+_6411636 | 0.38 |
ENST00000299397.3 ENST00000356761.2 ENST00000342245.4 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr16_+_53133070 | 0.35 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr12_-_76879852 | 0.34 |
ENST00000548341.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr3_+_69928256 | 0.33 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr10_-_49860525 | 0.33 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr7_+_150065278 | 0.31 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr5_+_140602904 | 0.31 |
ENST00000515856.2 ENST00000239449.4 |
PCDHB14 |
protocadherin beta 14 |
| chr8_+_22424551 | 0.31 |
ENST00000523348.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr5_+_76011868 | 0.31 |
ENST00000319211.4 |
F2R |
coagulation factor II (thrombin) receptor |
| chr5_+_67586465 | 0.30 |
ENST00000336483.5 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr8_+_104383728 | 0.28 |
ENST00000330295.5 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr4_-_138453606 | 0.28 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr3_+_69812701 | 0.28 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr1_+_40713573 | 0.27 |
ENST00000372766.3 |
TMCO2 |
transmembrane and coiled-coil domains 2 |
| chr16_-_3350614 | 0.27 |
ENST00000268674.2 |
TIGD7 |
tigger transposable element derived 7 |
| chr2_-_224467093 | 0.26 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr7_-_138363824 | 0.25 |
ENST00000419765.3 |
SVOPL |
SVOP-like |
| chrX_-_71351678 | 0.25 |
ENST00000609883.1 ENST00000545866.1 |
RGAG4 |
retrotransposon gag domain containing 4 |
| chr7_-_14026063 | 0.25 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr5_-_127674883 | 0.25 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr8_+_77318769 | 0.24 |
ENST00000518732.1 |
RP11-706J10.1 |
long intergenic non-protein coding RNA 1111 |
| chr2_+_109237717 | 0.24 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr6_-_154751629 | 0.24 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr1_+_144989309 | 0.24 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr12_-_91505608 | 0.24 |
ENST00000266718.4 |
LUM |
lumican |
| chrX_+_100333709 | 0.24 |
ENST00000372930.4 |
TMEM35 |
transmembrane protein 35 |
| chr20_+_11898507 | 0.23 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr20_+_12989596 | 0.23 |
ENST00000434210.1 ENST00000399002.2 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
| chr1_-_243326612 | 0.23 |
ENST00000492145.1 ENST00000490813.1 ENST00000464936.1 |
CEP170 |
centrosomal protein 170kDa |
| chr11_-_121986923 | 0.23 |
ENST00000560104.1 |
BLID |
BH3-like motif containing, cell death inducer |
| chr1_+_145439306 | 0.23 |
ENST00000425134.1 |
TXNIP |
thioredoxin interacting protein |
| chr5_+_140855495 | 0.23 |
ENST00000308177.3 |
PCDHGC3 |
protocadherin gamma subfamily C, 3 |
| chr8_-_122653630 | 0.23 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr6_+_64346386 | 0.22 |
ENST00000509330.1 |
PHF3 |
PHD finger protein 3 |
| chr6_+_28249299 | 0.22 |
ENST00000405948.2 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr11_+_71935797 | 0.22 |
ENST00000298229.2 ENST00000541756.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr2_+_158114051 | 0.22 |
ENST00000259056.4 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr12_+_104697504 | 0.21 |
ENST00000527879.1 |
EID3 |
EP300 interacting inhibitor of differentiation 3 |
| chr6_+_28249332 | 0.21 |
ENST00000259883.3 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr4_-_74853897 | 0.21 |
ENST00000296028.3 |
PPBP |
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr22_+_46476192 | 0.21 |
ENST00000443490.1 |
FLJ27365 |
hsa-mir-4763 |
| chr6_+_167704798 | 0.21 |
ENST00000230256.3 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr15_-_55657428 | 0.21 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr20_+_56964169 | 0.21 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr7_-_112635675 | 0.21 |
ENST00000447785.1 ENST00000451962.1 |
AC018464.3 |
AC018464.3 |
| chr5_-_133510456 | 0.21 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr6_-_29055090 | 0.20 |
ENST00000377173.2 |
OR2B3 |
olfactory receptor, family 2, subfamily B, member 3 |
| chr7_-_14026123 | 0.20 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr7_+_99717230 | 0.20 |
ENST00000262932.3 |
CNPY4 |
canopy FGF signaling regulator 4 |
| chr12_-_65146636 | 0.20 |
ENST00000418919.2 |
GNS |
glucosamine (N-acetyl)-6-sulfatase |
| chr6_+_151646800 | 0.20 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr6_+_167704838 | 0.20 |
ENST00000366829.2 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chrX_-_107975917 | 0.20 |
ENST00000563887.1 |
RP6-24A23.6 |
Uncharacterized protein |
| chr11_-_58612168 | 0.19 |
ENST00000287275.1 |
GLYATL2 |
glycine-N-acyltransferase-like 2 |
| chr4_+_70894130 | 0.19 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr7_-_92777606 | 0.19 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr6_+_153552455 | 0.19 |
ENST00000392385.2 |
AL590867.1 |
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr15_-_31393910 | 0.19 |
ENST00000397795.2 ENST00000256552.6 ENST00000559179.1 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
| chr5_-_140700322 | 0.19 |
ENST00000313368.5 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chrX_+_22050546 | 0.19 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr14_+_51955831 | 0.19 |
ENST00000356218.4 |
FRMD6 |
FERM domain containing 6 |
| chr7_-_14029283 | 0.19 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr5_-_150473127 | 0.19 |
ENST00000521001.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr13_+_24144509 | 0.18 |
ENST00000248484.4 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr17_-_53800217 | 0.18 |
ENST00000424486.2 |
TMEM100 |
transmembrane protein 100 |
| chr1_+_210501589 | 0.18 |
ENST00000413764.2 ENST00000541565.1 |
HHAT |
hedgehog acyltransferase |
| chr12_-_71551868 | 0.18 |
ENST00000247829.3 |
TSPAN8 |
tetraspanin 8 |
| chr1_-_150780757 | 0.18 |
ENST00000271651.3 |
CTSK |
cathepsin K |
| chr7_-_137028498 | 0.18 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr10_-_28571015 | 0.18 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr11_+_120973375 | 0.18 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr1_+_170501270 | 0.17 |
ENST00000367763.3 ENST00000367762.1 |
GORAB |
golgin, RAB6-interacting |
| chr7_+_120702819 | 0.17 |
ENST00000423795.1 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr10_-_14372870 | 0.17 |
ENST00000357447.2 |
FRMD4A |
FERM domain containing 4A |
| chr7_-_137028534 | 0.17 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr3_-_178984759 | 0.17 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr15_-_55700522 | 0.16 |
ENST00000564092.1 ENST00000310958.6 |
CCPG1 |
cell cycle progression 1 |
| chr19_-_40023450 | 0.16 |
ENST00000326282.4 |
EID2B |
EP300 interacting inhibitor of differentiation 2B |
| chr1_+_77747656 | 0.16 |
ENST00000354567.2 |
AK5 |
adenylate kinase 5 |
| chr10_-_50747064 | 0.16 |
ENST00000355832.5 ENST00000603152.1 ENST00000447839.2 |
ERCC6 PGBD3 ERCC6-PGBD3 |
excision repair cross-complementing rodent repair deficiency, complementation group 6 piggyBac transposable element derived 3 ERCC6-PGBD3 readthrough |
| chr7_-_14028488 | 0.16 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr4_+_62067860 | 0.16 |
ENST00000514591.1 |
LPHN3 |
latrophilin 3 |
| chr20_-_25371412 | 0.15 |
ENST00000339157.5 ENST00000376542.3 |
ABHD12 |
abhydrolase domain containing 12 |
| chr4_-_119759795 | 0.15 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr3_-_150690471 | 0.15 |
ENST00000468836.1 ENST00000328863.4 |
CLRN1 |
clarin 1 |
| chr20_-_30310656 | 0.15 |
ENST00000376055.4 |
BCL2L1 |
BCL2-like 1 |
| chr5_-_78281603 | 0.15 |
ENST00000264914.4 |
ARSB |
arylsulfatase B |
| chr5_+_118788182 | 0.15 |
ENST00000515320.1 ENST00000510025.1 |
HSD17B4 |
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr10_-_28270795 | 0.15 |
ENST00000545014.1 |
ARMC4 |
armadillo repeat containing 4 |
| chr4_-_165898768 | 0.15 |
ENST00000329314.5 |
TRIM61 |
tripartite motif containing 61 |
| chr20_-_30310336 | 0.14 |
ENST00000434194.1 ENST00000376062.2 |
BCL2L1 |
BCL2-like 1 |
| chr8_-_25281747 | 0.14 |
ENST00000421054.2 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr2_-_203735976 | 0.14 |
ENST00000435143.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr5_+_135496675 | 0.14 |
ENST00000507637.1 |
SMAD5 |
SMAD family member 5 |
| chr10_-_33623310 | 0.14 |
ENST00000395995.1 ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1 |
neuropilin 1 |
| chr9_+_2157655 | 0.14 |
ENST00000452193.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_-_10978957 | 0.14 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr21_+_31768348 | 0.14 |
ENST00000355459.2 |
KRTAP13-1 |
keratin associated protein 13-1 |
| chr13_+_49551020 | 0.14 |
ENST00000541916.1 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr3_-_149095652 | 0.14 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr11_-_33913708 | 0.13 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr15_+_55700741 | 0.13 |
ENST00000569691.1 |
C15orf65 |
chromosome 15 open reading frame 65 |
| chr16_+_8806800 | 0.13 |
ENST00000561870.1 ENST00000396600.2 |
ABAT |
4-aminobutyrate aminotransferase |
| chr12_-_10605929 | 0.13 |
ENST00000347831.5 ENST00000359151.3 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr17_-_39191107 | 0.13 |
ENST00000344363.5 |
KRTAP1-3 |
keratin associated protein 1-3 |
| chr2_-_203735484 | 0.13 |
ENST00000420558.1 ENST00000418208.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr8_+_24298531 | 0.13 |
ENST00000175238.6 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chrX_-_101410762 | 0.13 |
ENST00000543160.1 ENST00000333643.3 |
BEX5 |
brain expressed, X-linked 5 |
| chr1_+_115572415 | 0.13 |
ENST00000256592.1 |
TSHB |
thyroid stimulating hormone, beta |
| chr5_+_141348640 | 0.12 |
ENST00000540015.1 ENST00000506938.1 ENST00000394514.2 ENST00000512565.1 ENST00000394515.3 |
RNF14 |
ring finger protein 14 |
| chr6_+_34204642 | 0.12 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr5_-_160279207 | 0.12 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr6_+_73076432 | 0.12 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr2_-_203735586 | 0.12 |
ENST00000454326.1 ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr12_+_15699286 | 0.12 |
ENST00000442921.2 ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr5_+_140710061 | 0.12 |
ENST00000517417.1 ENST00000378105.3 |
PCDHGA1 |
protocadherin gamma subfamily A, 1 |
| chr7_-_14029515 | 0.12 |
ENST00000430479.1 ENST00000405218.2 ENST00000343495.5 |
ETV1 |
ets variant 1 |
| chr1_+_153190053 | 0.12 |
ENST00000368744.3 |
PRR9 |
proline rich 9 |
| chr11_+_77300669 | 0.12 |
ENST00000313578.3 |
AQP11 |
aquaporin 11 |
| chr11_-_124981475 | 0.12 |
ENST00000532156.1 ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218 |
transmembrane protein 218 |
| chr7_+_142458507 | 0.12 |
ENST00000492062.1 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
| chr4_-_83765613 | 0.12 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr2_-_225266743 | 0.11 |
ENST00000409685.3 |
FAM124B |
family with sequence similarity 124B |
| chr11_-_26593677 | 0.11 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr1_-_157746909 | 0.11 |
ENST00000392274.3 ENST00000361516.3 ENST00000368181.4 |
FCRL2 |
Fc receptor-like 2 |
| chr5_-_148929848 | 0.11 |
ENST00000504676.1 ENST00000515435.1 |
CSNK1A1 |
casein kinase 1, alpha 1 |
| chr1_+_225600404 | 0.11 |
ENST00000366845.2 |
AC092811.1 |
AC092811.1 |
| chr10_-_14996321 | 0.11 |
ENST00000378289.4 |
DCLRE1C |
DNA cross-link repair 1C |
| chr13_-_41593425 | 0.11 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chrX_+_36065053 | 0.11 |
ENST00000313548.4 |
CHDC2 |
calponin homology domain containing 2 |
| chr22_+_31160239 | 0.11 |
ENST00000445781.1 ENST00000401475.1 |
OSBP2 |
oxysterol binding protein 2 |
| chr20_-_13971255 | 0.11 |
ENST00000284951.5 ENST00000378072.5 |
SEL1L2 |
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr7_-_92855762 | 0.11 |
ENST00000453812.2 ENST00000394468.2 |
HEPACAM2 |
HEPACAM family member 2 |
| chr19_-_10445399 | 0.11 |
ENST00000592945.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr17_+_47448102 | 0.11 |
ENST00000576461.1 |
RP11-81K2.1 |
Uncharacterized protein |
| chrX_+_36246735 | 0.11 |
ENST00000378653.3 |
CXorf30 |
chromosome X open reading frame 30 |
| chr6_+_140175987 | 0.11 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr8_+_105352050 | 0.11 |
ENST00000297581.2 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
| chr1_+_152943122 | 0.11 |
ENST00000328051.2 |
SPRR4 |
small proline-rich protein 4 |
| chr12_-_103310987 | 0.11 |
ENST00000307000.2 |
PAH |
phenylalanine hydroxylase |
| chr5_+_54455946 | 0.11 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr1_+_104159999 | 0.10 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr12_-_53171128 | 0.10 |
ENST00000332411.2 |
KRT76 |
keratin 76 |
| chr15_+_76629064 | 0.10 |
ENST00000290759.4 |
ISL2 |
ISL LIM homeobox 2 |
| chr3_-_55001115 | 0.10 |
ENST00000493075.1 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
| chr1_+_43291220 | 0.10 |
ENST00000372514.3 |
ERMAP |
erythroblast membrane-associated protein (Scianna blood group) |
| chr2_+_45168875 | 0.10 |
ENST00000260653.3 |
SIX3 |
SIX homeobox 3 |
| chrX_-_18690210 | 0.10 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
| chr5_+_108083517 | 0.10 |
ENST00000281092.4 ENST00000536402.1 |
FER |
fer (fps/fes related) tyrosine kinase |
| chr12_+_56367697 | 0.10 |
ENST00000553116.1 ENST00000360299.5 ENST00000548068.1 ENST00000549915.1 ENST00000551459.1 ENST00000448789.2 |
RAB5B |
RAB5B, member RAS oncogene family |
| chr18_+_29171689 | 0.10 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr4_-_70725856 | 0.10 |
ENST00000226444.3 |
SULT1E1 |
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr8_+_38831683 | 0.10 |
ENST00000302495.4 |
HTRA4 |
HtrA serine peptidase 4 |
| chr11_-_17229480 | 0.10 |
ENST00000532035.1 ENST00000540361.1 |
PIK3C2A |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr3_-_178976996 | 0.10 |
ENST00000485523.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chrX_+_139791917 | 0.10 |
ENST00000607004.1 ENST00000370535.3 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
| chr19_+_15852203 | 0.10 |
ENST00000305892.1 |
OR10H3 |
olfactory receptor, family 10, subfamily H, member 3 |
| chr7_-_120498357 | 0.10 |
ENST00000415871.1 ENST00000222747.3 ENST00000430985.1 |
TSPAN12 |
tetraspanin 12 |
| chr8_+_24298597 | 0.10 |
ENST00000380789.1 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr2_-_188312971 | 0.10 |
ENST00000410068.1 ENST00000447403.1 ENST00000410102.1 |
CALCRL |
calcitonin receptor-like |
| chr5_-_125930929 | 0.10 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr1_-_182361327 | 0.10 |
ENST00000331872.6 ENST00000311223.5 |
GLUL |
glutamate-ammonia ligase |
| chrX_-_153602991 | 0.10 |
ENST00000369850.3 ENST00000422373.1 |
FLNA |
filamin A, alpha |
| chr1_+_12851545 | 0.10 |
ENST00000332296.7 |
PRAMEF1 |
PRAME family member 1 |
| chr21_-_31869451 | 0.10 |
ENST00000334058.2 |
KRTAP19-4 |
keratin associated protein 19-4 |
| chr1_-_190446759 | 0.10 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr7_-_2883928 | 0.09 |
ENST00000275364.3 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
| chr12_-_71551652 | 0.09 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.0 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.2 | 0.3 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 1.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.8 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 4.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.2 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.0 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0009183 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.0 | 0.1 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.8 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:1905040 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) positive regulation of calcium-transporting ATPase activity(GO:1901896) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0030324 | lung development(GO:0030324) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 3.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 1.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 6.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 5.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0030695 | GTPase regulator activity(GO:0030695) nucleoside-triphosphatase regulator activity(GO:0060589) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 4.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |