Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for BHLHE40
Z-value: 0.63
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.4 | BHLHE40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE40 | hg19_v2_chr3_+_5020801_5020952 | -0.30 | 4.7e-01 | Click! |
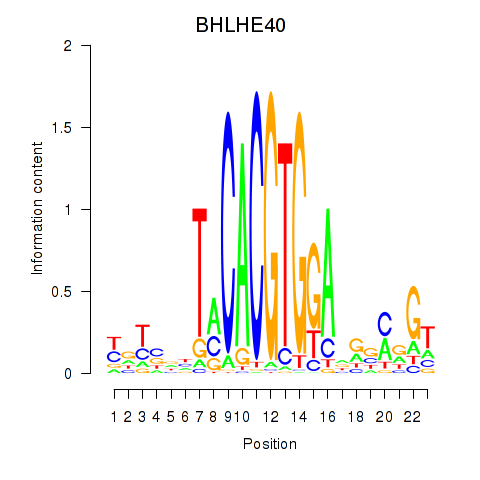
Activity profile of BHLHE40 motif
Sorted Z-values of BHLHE40 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE40
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_136871957 | 0.85 |
ENST00000354570.3 |
MAP7 |
microtubule-associated protein 7 |
| chr1_+_35247859 | 0.76 |
ENST00000373362.3 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr17_-_17109579 | 0.66 |
ENST00000321560.3 |
PLD6 |
phospholipase D family, member 6 |
| chr7_+_147830776 | 0.56 |
ENST00000538075.1 |
CNTNAP2 |
contactin associated protein-like 2 |
| chr8_+_32405785 | 0.53 |
ENST00000287842.3 |
NRG1 |
neuregulin 1 |
| chr8_+_32405728 | 0.51 |
ENST00000523079.1 ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1 |
neuregulin 1 |
| chr10_-_6019552 | 0.37 |
ENST00000379977.3 ENST00000397251.3 ENST00000397248.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr19_-_16045665 | 0.35 |
ENST00000248041.8 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr19_-_16045619 | 0.35 |
ENST00000402119.4 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr12_-_48298785 | 0.34 |
ENST00000550325.1 ENST00000546653.1 ENST00000549336.1 ENST00000535672.1 ENST00000229022.3 ENST00000548664.1 |
VDR |
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr9_+_706842 | 0.33 |
ENST00000382293.3 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr12_-_25101920 | 0.32 |
ENST00000539780.1 ENST00000546285.1 ENST00000342945.5 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr5_+_89770664 | 0.32 |
ENST00000503973.1 ENST00000399107.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr5_+_89770696 | 0.30 |
ENST00000504930.1 ENST00000514483.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr11_+_60609537 | 0.30 |
ENST00000227520.5 |
CCDC86 |
coiled-coil domain containing 86 |
| chr19_-_55652290 | 0.29 |
ENST00000589745.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr8_-_72459885 | 0.27 |
ENST00000523987.1 |
RP11-1102P16.1 |
Uncharacterized protein |
| chr8_+_99129513 | 0.26 |
ENST00000522319.1 ENST00000401707.2 |
POP1 |
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr9_-_117111222 | 0.23 |
ENST00000374079.4 |
AKNA |
AT-hook transcription factor |
| chr1_+_210001309 | 0.23 |
ENST00000491415.2 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
| chr5_+_149737202 | 0.22 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
| chr14_+_77924373 | 0.22 |
ENST00000216479.3 ENST00000535854.2 ENST00000555517.1 |
AHSA1 |
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr2_-_136743169 | 0.21 |
ENST00000264161.4 |
DARS |
aspartyl-tRNA synthetase |
| chr14_-_102026643 | 0.21 |
ENST00000555882.1 ENST00000554441.1 ENST00000553729.1 ENST00000557109.1 ENST00000557532.1 ENST00000554694.1 ENST00000554735.1 ENST00000555174.1 ENST00000557661.1 |
DIO3OS |
DIO3 opposite strand/antisense RNA (head to head) |
| chr12_-_25102252 | 0.21 |
ENST00000261192.7 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr12_-_6677422 | 0.20 |
ENST00000382421.3 ENST00000545200.1 ENST00000399466.2 ENST00000536124.1 ENST00000540228.1 ENST00000542867.1 ENST00000545492.1 ENST00000322166.5 ENST00000545915.1 |
NOP2 |
NOP2 nucleolar protein |
| chr1_+_9648921 | 0.20 |
ENST00000377376.4 ENST00000340305.5 ENST00000340381.6 |
TMEM201 |
transmembrane protein 201 |
| chr1_+_173793777 | 0.19 |
ENST00000239457.5 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr19_+_35645618 | 0.19 |
ENST00000392218.2 ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
| chr2_-_136743039 | 0.19 |
ENST00000537273.1 |
DARS |
aspartyl-tRNA synthetase |
| chrX_+_54556633 | 0.18 |
ENST00000336470.4 ENST00000360845.2 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr9_+_130565147 | 0.18 |
ENST00000373247.2 ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS |
folylpolyglutamate synthase |
| chr1_-_43637915 | 0.17 |
ENST00000236051.2 |
EBNA1BP2 |
EBNA1 binding protein 2 |
| chr7_-_37024665 | 0.16 |
ENST00000396040.2 |
ELMO1 |
engulfment and cell motility 1 |
| chr2_+_207630081 | 0.16 |
ENST00000236980.6 ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2 |
FAST kinase domains 2 |
| chr6_+_144164455 | 0.16 |
ENST00000367576.5 |
LTV1 |
LTV1 homolog (S. cerevisiae) |
| chr17_+_33307503 | 0.16 |
ENST00000378526.4 ENST00000585941.1 ENST00000262327.5 ENST00000592690.1 ENST00000585740.1 |
LIG3 |
ligase III, DNA, ATP-dependent |
| chr14_+_73393040 | 0.15 |
ENST00000358377.2 ENST00000353777.3 ENST00000394234.2 ENST00000509153.1 ENST00000555042.1 |
DCAF4 |
DDB1 and CUL4 associated factor 4 |
| chr1_-_43638168 | 0.15 |
ENST00000431635.2 |
EBNA1BP2 |
EBNA1 binding protein 2 |
| chr2_+_103236004 | 0.15 |
ENST00000233969.2 |
SLC9A2 |
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr1_-_200589859 | 0.15 |
ENST00000367350.4 |
KIF14 |
kinesin family member 14 |
| chrX_+_129040122 | 0.15 |
ENST00000394422.3 ENST00000371051.5 |
UTP14A |
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr10_-_6019455 | 0.15 |
ENST00000530685.1 ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr1_-_113498943 | 0.14 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr12_-_55042140 | 0.14 |
ENST00000293371.6 ENST00000456047.2 |
DCD |
dermcidin |
| chr17_-_13505219 | 0.14 |
ENST00000284110.1 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr2_-_112642267 | 0.13 |
ENST00000341068.3 |
ANAPC1 |
anaphase promoting complex subunit 1 |
| chr21_+_33784670 | 0.13 |
ENST00000300255.2 |
EVA1C |
eva-1 homolog C (C. elegans) |
| chr8_+_109455845 | 0.13 |
ENST00000220853.3 |
EMC2 |
ER membrane protein complex subunit 2 |
| chr15_+_68924327 | 0.13 |
ENST00000543950.1 |
CORO2B |
coronin, actin binding protein, 2B |
| chr13_-_99630233 | 0.13 |
ENST00000376460.1 ENST00000442173.1 |
DOCK9 |
dedicator of cytokinesis 9 |
| chrX_+_129040094 | 0.13 |
ENST00000425117.2 |
UTP14A |
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr6_-_154677900 | 0.13 |
ENST00000265198.4 ENST00000520261.1 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
| chr19_+_35645817 | 0.13 |
ENST00000423817.3 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
| chr1_-_112046289 | 0.13 |
ENST00000241356.4 |
ADORA3 |
adenosine A3 receptor |
| chr4_+_47487285 | 0.12 |
ENST00000273859.3 ENST00000504445.1 |
ATP10D |
ATPase, class V, type 10D |
| chr5_-_101834617 | 0.12 |
ENST00000513675.1 ENST00000379807.3 |
SLCO6A1 |
solute carrier organic anion transporter family, member 6A1 |
| chr15_-_74988281 | 0.12 |
ENST00000566828.1 ENST00000563009.1 ENST00000568176.1 ENST00000566243.1 ENST00000566219.1 ENST00000426797.3 ENST00000566119.1 ENST00000315127.4 |
EDC3 |
enhancer of mRNA decapping 3 |
| chr19_+_7660716 | 0.12 |
ENST00000160298.4 ENST00000446248.2 |
CAMSAP3 |
calmodulin regulated spectrin-associated protein family, member 3 |
| chr11_+_34664014 | 0.12 |
ENST00000527935.1 |
EHF |
ets homologous factor |
| chr8_-_11710979 | 0.12 |
ENST00000415599.2 |
CTSB |
cathepsin B |
| chr19_-_5719860 | 0.12 |
ENST00000590729.1 |
LONP1 |
lon peptidase 1, mitochondrial |
| chr12_-_58165870 | 0.11 |
ENST00000257848.7 |
METTL1 |
methyltransferase like 1 |
| chr8_-_134584092 | 0.11 |
ENST00000522652.1 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr1_-_207095324 | 0.11 |
ENST00000530505.1 ENST00000367091.3 ENST00000442471.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr8_-_48872686 | 0.11 |
ENST00000314191.2 ENST00000338368.3 |
PRKDC |
protein kinase, DNA-activated, catalytic polypeptide |
| chr22_-_30987849 | 0.11 |
ENST00000402284.3 ENST00000354694.7 |
PES1 |
pescadillo ribosomal biogenesis factor 1 |
| chr22_-_30987837 | 0.10 |
ENST00000335214.6 |
PES1 |
pescadillo ribosomal biogenesis factor 1 |
| chr4_-_57301748 | 0.10 |
ENST00000264220.2 |
PPAT |
phosphoribosyl pyrophosphate amidotransferase |
| chr1_+_207070775 | 0.10 |
ENST00000391929.3 ENST00000294984.2 ENST00000367093.3 |
IL24 |
interleukin 24 |
| chr8_+_22857048 | 0.10 |
ENST00000251822.6 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
| chr7_-_138720763 | 0.10 |
ENST00000275766.1 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
| chr18_+_580367 | 0.10 |
ENST00000327228.3 |
CETN1 |
centrin, EF-hand protein, 1 |
| chr2_+_183989157 | 0.10 |
ENST00000541912.1 |
NUP35 |
nucleoporin 35kDa |
| chr1_-_171621815 | 0.10 |
ENST00000037502.6 |
MYOC |
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr5_-_101834712 | 0.09 |
ENST00000506729.1 ENST00000389019.3 ENST00000379810.1 |
SLCO6A1 |
solute carrier organic anion transporter family, member 6A1 |
| chr2_+_183989083 | 0.09 |
ENST00000295119.4 |
NUP35 |
nucleoporin 35kDa |
| chr1_+_118472343 | 0.09 |
ENST00000369441.3 ENST00000349139.5 |
WDR3 |
WD repeat domain 3 |
| chr14_+_22694606 | 0.09 |
ENST00000390463.3 |
TRAV36DV7 |
T cell receptor alpha variable 36/delta variable 7 |
| chr19_-_3786253 | 0.09 |
ENST00000585778.1 |
MATK |
megakaryocyte-associated tyrosine kinase |
| chr12_-_113772835 | 0.09 |
ENST00000552014.1 ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr19_-_45909585 | 0.09 |
ENST00000593226.1 ENST00000418234.2 |
PPP1R13L |
protein phosphatase 1, regulatory subunit 13 like |
| chr17_-_74733404 | 0.08 |
ENST00000508921.3 ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2 |
serine/arginine-rich splicing factor 2 |
| chr3_+_63638339 | 0.08 |
ENST00000343837.3 ENST00000469440.1 |
SNTN |
sentan, cilia apical structure protein |
| chr21_+_27107672 | 0.08 |
ENST00000400075.3 |
GABPA |
GA binding protein transcription factor, alpha subunit 60kDa |
| chr3_-_98312548 | 0.07 |
ENST00000264193.2 |
CPOX |
coproporphyrinogen oxidase |
| chr1_-_165325939 | 0.07 |
ENST00000342310.3 |
LMX1A |
LIM homeobox transcription factor 1, alpha |
| chr6_-_109804412 | 0.07 |
ENST00000230122.3 |
ZBTB24 |
zinc finger and BTB domain containing 24 |
| chr14_-_45722605 | 0.07 |
ENST00000310806.4 |
MIS18BP1 |
MIS18 binding protein 1 |
| chr5_+_36152091 | 0.07 |
ENST00000274254.5 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr1_+_12079517 | 0.07 |
ENST00000235332.4 ENST00000436478.2 |
MIIP |
migration and invasion inhibitory protein |
| chr6_+_5261225 | 0.07 |
ENST00000324331.6 |
FARS2 |
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr17_-_61850894 | 0.06 |
ENST00000403162.3 ENST00000582252.1 ENST00000225726.5 |
CCDC47 |
coiled-coil domain containing 47 |
| chr17_-_10600818 | 0.06 |
ENST00000577427.1 ENST00000255390.5 |
SCO1 |
SCO1 cytochrome c oxidase assembly protein |
| chr6_+_31865552 | 0.06 |
ENST00000469372.1 ENST00000497706.1 |
C2 |
complement component 2 |
| chr15_-_23692381 | 0.06 |
ENST00000567107.1 ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2 |
golgin A6 family-like 2 |
| chr22_-_38245304 | 0.06 |
ENST00000609454.1 |
ANKRD54 |
ankyrin repeat domain 54 |
| chrX_-_128977364 | 0.06 |
ENST00000371064.3 |
ZDHHC9 |
zinc finger, DHHC-type containing 9 |
| chr17_+_72772621 | 0.06 |
ENST00000335464.5 ENST00000417024.2 ENST00000578764.1 ENST00000582773.1 ENST00000582330.1 |
TMEM104 |
transmembrane protein 104 |
| chr8_+_125985531 | 0.06 |
ENST00000319286.5 |
ZNF572 |
zinc finger protein 572 |
| chr4_+_57302297 | 0.06 |
ENST00000399688.3 ENST00000512576.1 |
PAICS |
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr4_+_57301896 | 0.06 |
ENST00000514888.1 ENST00000264221.2 ENST00000505164.1 |
PAICS |
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr4_-_85418103 | 0.06 |
ENST00000515820.2 |
NKX6-1 |
NK6 homeobox 1 |
| chr5_-_89770582 | 0.06 |
ENST00000316610.6 |
MBLAC2 |
metallo-beta-lactamase domain containing 2 |
| chr10_+_101491968 | 0.05 |
ENST00000370476.5 ENST00000370472.4 |
CUTC |
cutC copper transporter |
| chrX_-_128977781 | 0.05 |
ENST00000357166.6 |
ZDHHC9 |
zinc finger, DHHC-type containing 9 |
| chr22_-_50523760 | 0.05 |
ENST00000395876.2 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr6_+_44215603 | 0.05 |
ENST00000371554.1 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr7_-_151574191 | 0.05 |
ENST00000287878.4 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr21_+_38071430 | 0.05 |
ENST00000290399.6 |
SIM2 |
single-minded family bHLH transcription factor 2 |
| chr9_-_95640218 | 0.05 |
ENST00000395506.3 ENST00000375495.3 ENST00000332591.6 |
ZNF484 |
zinc finger protein 484 |
| chr7_+_2671568 | 0.05 |
ENST00000258796.7 |
TTYH3 |
tweety family member 3 |
| chr5_+_36152163 | 0.04 |
ENST00000274255.6 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr7_-_155326532 | 0.04 |
ENST00000406197.1 ENST00000321736.5 |
CNPY1 |
canopy FGF signaling regulator 1 |
| chr10_-_35379241 | 0.04 |
ENST00000374748.1 ENST00000374749.3 |
CUL2 |
cullin 2 |
| chr6_+_13615554 | 0.04 |
ENST00000451315.2 |
NOL7 |
nucleolar protein 7, 27kDa |
| chr19_+_13106383 | 0.04 |
ENST00000397661.2 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr12_+_93861282 | 0.04 |
ENST00000552217.1 ENST00000393128.4 ENST00000547098.1 |
MRPL42 |
mitochondrial ribosomal protein L42 |
| chr3_+_133293278 | 0.04 |
ENST00000508481.1 ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr12_+_93861264 | 0.04 |
ENST00000549982.1 ENST00000361630.2 |
MRPL42 |
mitochondrial ribosomal protein L42 |
| chr10_-_35379524 | 0.03 |
ENST00000374751.3 ENST00000374742.1 ENST00000602371.1 |
CUL2 |
cullin 2 |
| chr3_+_111578640 | 0.03 |
ENST00000393925.3 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
| chr9_-_123691047 | 0.03 |
ENST00000373887.3 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr4_+_41983713 | 0.03 |
ENST00000333141.5 |
DCAF4L1 |
DDB1 and CUL4 associated factor 4-like 1 |
| chr1_-_86174065 | 0.03 |
ENST00000370574.3 ENST00000431532.2 |
ZNHIT6 |
zinc finger, HIT-type containing 6 |
| chr17_+_66624280 | 0.03 |
ENST00000585484.1 |
RP11-118B18.1 |
RP11-118B18.1 |
| chr6_+_70576457 | 0.03 |
ENST00000322773.4 |
COL19A1 |
collagen, type XIX, alpha 1 |
| chr1_-_231376836 | 0.03 |
ENST00000451322.1 |
C1orf131 |
chromosome 1 open reading frame 131 |
| chr8_+_55047763 | 0.03 |
ENST00000260102.4 ENST00000519831.1 |
MRPL15 |
mitochondrial ribosomal protein L15 |
| chr11_-_6624801 | 0.03 |
ENST00000534343.1 ENST00000254605.6 |
RRP8 |
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr1_-_231376867 | 0.02 |
ENST00000366649.2 ENST00000318906.2 ENST00000366651.3 |
C1orf131 |
chromosome 1 open reading frame 131 |
| chr2_-_10587897 | 0.02 |
ENST00000405333.1 ENST00000443218.1 |
ODC1 |
ornithine decarboxylase 1 |
| chr17_-_7193711 | 0.02 |
ENST00000571464.1 |
YBX2 |
Y box binding protein 2 |
| chr5_+_36152179 | 0.02 |
ENST00000508514.1 ENST00000513151.1 ENST00000546211.1 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chrX_+_23685563 | 0.02 |
ENST00000379341.4 |
PRDX4 |
peroxiredoxin 4 |
| chr2_+_105654441 | 0.02 |
ENST00000258455.3 |
MRPS9 |
mitochondrial ribosomal protein S9 |
| chr2_-_241497390 | 0.01 |
ENST00000272972.3 ENST00000401804.1 ENST00000361678.4 ENST00000405523.3 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
| chr9_-_100881466 | 0.01 |
ENST00000341469.2 ENST00000342043.3 ENST00000375098.3 |
TRIM14 |
tripartite motif containing 14 |
| chr17_+_15635561 | 0.01 |
ENST00000584301.1 ENST00000580596.1 ENST00000464963.1 ENST00000437605.2 ENST00000579428.1 |
TBC1D26 |
TBC1 domain family, member 26 |
| chr2_-_10588630 | 0.01 |
ENST00000234111.4 |
ODC1 |
ornithine decarboxylase 1 |
| chr17_+_74733744 | 0.01 |
ENST00000586689.1 ENST00000587661.1 ENST00000593181.1 ENST00000336509.4 ENST00000355954.3 |
MFSD11 |
major facilitator superfamily domain containing 11 |
| chr1_+_119957554 | 0.01 |
ENST00000543831.1 ENST00000433745.1 ENST00000369416.3 |
HSD3B2 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr16_+_4897912 | 0.01 |
ENST00000545171.1 |
UBN1 |
ubinuclein 1 |
| chr22_+_48885272 | 0.01 |
ENST00000402357.1 |
FAM19A5 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chrX_+_23685653 | 0.01 |
ENST00000379331.3 |
PRDX4 |
peroxiredoxin 4 |
| chr22_+_22681656 | 0.01 |
ENST00000390291.2 |
IGLV1-50 |
immunoglobulin lambda variable 1-50 (non-functional) |
| chr19_+_41497178 | 0.01 |
ENST00000324071.4 |
CYP2B6 |
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr2_+_127413481 | 0.01 |
ENST00000259254.4 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr17_-_72772462 | 0.01 |
ENST00000582870.1 ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9 |
N-acetyltransferase 9 (GCN5-related, putative) |
| chr8_-_134584152 | 0.00 |
ENST00000521180.1 ENST00000517668.1 ENST00000319914.5 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr2_-_47143160 | 0.00 |
ENST00000409800.1 ENST00000409218.1 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr19_+_15121532 | 0.00 |
ENST00000292574.3 |
CCDC105 |
coiled-coil domain containing 105 |
| chr1_+_76262552 | 0.00 |
ENST00000263187.3 |
MSH4 |
mutS homolog 4 |
| chr10_-_21435488 | 0.00 |
ENST00000534331.1 ENST00000529198.1 ENST00000377118.4 |
C10orf113 |
chromosome 10 open reading frame 113 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 0.7 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 | 1.0 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.3 | GO:1902339 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.3 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.1 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.5 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.2 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.1 | 0.7 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.3 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0005694 | chromosome(GO:0005694) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 1.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.5 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0038181 | bile acid receptor activity(GO:0038181) vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0070364 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |