Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
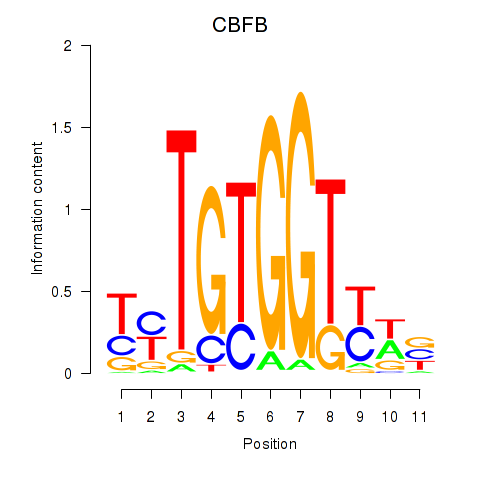
Results for CBFB
Z-value: 0.39
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | CBFB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg19_v2_chr16_+_67063142_67063153 | -0.33 | 4.2e-01 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_76210448 | 0.39 |
ENST00000377499.5 |
LMO7 |
LIM domain 7 |
| chr2_-_158300556 | 0.39 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr3_+_157154578 | 0.37 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr14_-_61116168 | 0.36 |
ENST00000247182.6 |
SIX1 |
SIX homeobox 1 |
| chr8_+_104384616 | 0.21 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr8_-_60031762 | 0.19 |
ENST00000361421.1 |
TOX |
thymocyte selection-associated high mobility group box |
| chr17_+_1633755 | 0.19 |
ENST00000545662.1 |
WDR81 |
WD repeat domain 81 |
| chr7_+_120628731 | 0.17 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr14_+_85996507 | 0.16 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr9_-_130541017 | 0.15 |
ENST00000314830.8 |
SH2D3C |
SH2 domain containing 3C |
| chr6_-_75912508 | 0.13 |
ENST00000416123.2 |
COL12A1 |
collagen, type XII, alpha 1 |
| chr14_+_85996471 | 0.12 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_39876151 | 0.11 |
ENST00000530275.1 |
KIAA0754 |
KIAA0754 |
| chr3_-_71834318 | 0.10 |
ENST00000353065.3 |
PROK2 |
prokineticin 2 |
| chr11_-_9781068 | 0.09 |
ENST00000500698.1 |
RP11-540A21.2 |
RP11-540A21.2 |
| chr9_-_123342415 | 0.09 |
ENST00000349780.4 ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr17_+_32582293 | 0.09 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr1_+_17634689 | 0.08 |
ENST00000375453.1 ENST00000375448.4 |
PADI4 |
peptidyl arginine deiminase, type IV |
| chr19_-_14048804 | 0.08 |
ENST00000254320.3 ENST00000586075.1 |
PODNL1 |
podocan-like 1 |
| chr22_+_23222886 | 0.08 |
ENST00000390319.2 |
IGLV3-1 |
immunoglobulin lambda variable 3-1 |
| chr19_-_14049184 | 0.08 |
ENST00000339560.5 |
PODNL1 |
podocan-like 1 |
| chr21_-_36421626 | 0.08 |
ENST00000300305.3 |
RUNX1 |
runt-related transcription factor 1 |
| chr14_-_85996332 | 0.08 |
ENST00000380722.1 |
RP11-497E19.1 |
RP11-497E19.1 |
| chr16_+_53088885 | 0.07 |
ENST00000566029.1 ENST00000447540.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chrX_-_19688475 | 0.07 |
ENST00000541422.1 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr12_-_53594227 | 0.07 |
ENST00000550743.2 |
ITGB7 |
integrin, beta 7 |
| chr21_-_36421535 | 0.06 |
ENST00000416754.1 ENST00000437180.1 ENST00000455571.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr1_-_179112173 | 0.06 |
ENST00000408940.3 ENST00000504405.1 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr6_+_30908747 | 0.06 |
ENST00000462446.1 ENST00000304311.2 |
DPCR1 |
diffuse panbronchiolitis critical region 1 |
| chr1_-_184006829 | 0.06 |
ENST00000361927.4 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
| chr6_+_27925019 | 0.05 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr11_-_66104237 | 0.05 |
ENST00000530056.1 |
RIN1 |
Ras and Rab interactor 1 |
| chr22_+_42665742 | 0.05 |
ENST00000332965.3 ENST00000415205.1 ENST00000446578.1 |
Z83851.3 |
Z83851.3 |
| chr1_+_89246647 | 0.05 |
ENST00000544045.1 |
PKN2 |
protein kinase N2 |
| chrX_-_153718988 | 0.05 |
ENST00000263512.4 ENST00000393587.4 ENST00000453912.1 |
SLC10A3 |
solute carrier family 10, member 3 |
| chr6_-_34524093 | 0.04 |
ENST00000544425.1 |
SPDEF |
SAM pointed domain containing ETS transcription factor |
| chr20_+_43343886 | 0.04 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr2_-_207024233 | 0.04 |
ENST00000423725.1 ENST00000233190.6 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr10_-_29811456 | 0.04 |
ENST00000535393.1 |
SVIL |
supervillin |
| chr11_-_66103867 | 0.03 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chrX_+_115567767 | 0.03 |
ENST00000371900.4 |
SLC6A14 |
solute carrier family 6 (amino acid transporter), member 14 |
| chr14_-_35183886 | 0.03 |
ENST00000298159.6 |
CFL2 |
cofilin 2 (muscle) |
| chr12_-_14849470 | 0.03 |
ENST00000261170.3 |
GUCY2C |
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr2_-_127977654 | 0.03 |
ENST00000409327.1 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr5_-_39203093 | 0.03 |
ENST00000515010.1 |
FYB |
FYN binding protein |
| chr13_-_28674693 | 0.03 |
ENST00000537084.1 ENST00000241453.7 ENST00000380982.4 |
FLT3 |
fms-related tyrosine kinase 3 |
| chr12_-_55028664 | 0.03 |
ENST00000547511.1 ENST00000257867.4 |
LACRT |
lacritin |
| chr1_-_184723942 | 0.03 |
ENST00000318130.8 |
EDEM3 |
ER degradation enhancer, mannosidase alpha-like 3 |
| chr11_-_66103932 | 0.03 |
ENST00000311320.4 |
RIN1 |
Ras and Rab interactor 1 |
| chr14_-_59951112 | 0.03 |
ENST00000247194.4 |
L3HYPDH |
L-3-hydroxyproline dehydratase (trans-) |
| chr2_+_68592305 | 0.03 |
ENST00000234313.7 |
PLEK |
pleckstrin |
| chr14_+_22748980 | 0.02 |
ENST00000390465.2 |
TRAV38-2DV8 |
T cell receptor alpha variable 38-2/delta variable 8 |
| chr19_+_10541462 | 0.02 |
ENST00000293683.5 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr6_+_52051171 | 0.02 |
ENST00000340057.1 |
IL17A |
interleukin 17A |
| chr1_-_240775447 | 0.02 |
ENST00000318160.4 |
GREM2 |
gremlin 2, DAN family BMP antagonist |
| chr5_-_108745689 | 0.02 |
ENST00000361189.2 |
PJA2 |
praja ring finger 2, E3 ubiquitin protein ligase |
| chr11_-_58343319 | 0.01 |
ENST00000395074.2 |
LPXN |
leupaxin |
| chr9_-_35080013 | 0.01 |
ENST00000378643.3 |
FANCG |
Fanconi anemia, complementation group G |
| chr4_-_156297949 | 0.01 |
ENST00000515654.1 |
MAP9 |
microtubule-associated protein 9 |
| chr12_+_15125954 | 0.01 |
ENST00000266395.2 |
PDE6H |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr12_-_52585765 | 0.01 |
ENST00000313234.5 ENST00000394815.2 |
KRT80 |
keratin 80 |
| chr8_+_28747884 | 0.01 |
ENST00000287701.10 ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1 |
homeobox containing 1 |
| chr6_-_34524049 | 0.01 |
ENST00000374037.3 |
SPDEF |
SAM pointed domain containing ETS transcription factor |
| chr1_+_196621002 | 0.01 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr17_-_39183452 | 0.00 |
ENST00000361883.5 |
KRTAP1-5 |
keratin associated protein 1-5 |
| chr14_+_72064945 | 0.00 |
ENST00000537413.1 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr12_-_4754339 | 0.00 |
ENST00000228850.1 |
AKAP3 |
A kinase (PRKA) anchor protein 3 |
| chr20_+_54987168 | 0.00 |
ENST00000360314.3 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr20_+_54987305 | 0.00 |
ENST00000371336.3 ENST00000434344.1 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr14_-_24977457 | 0.00 |
ENST00000250378.3 ENST00000206446.4 |
CMA1 |
chymase 1, mast cell |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.4 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |