Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
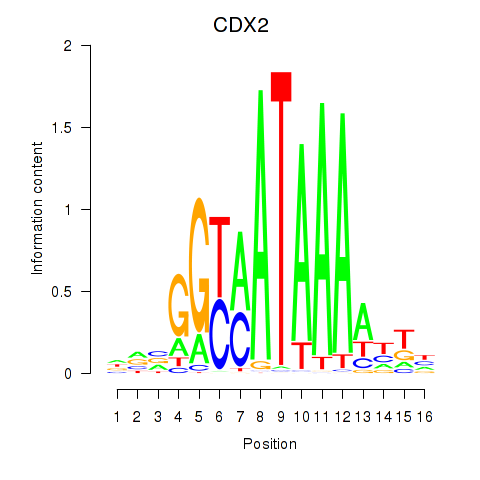
Results for CDX2
Z-value: 0.84
Transcription factors associated with CDX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX2
|
ENSG00000165556.9 | CDX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX2 | hg19_v2_chr13_-_28545276_28545276 | 0.36 | 3.7e-01 | Click! |
Activity profile of CDX2 motif
Sorted Z-values of CDX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_15939963 | 1.80 |
ENST00000259988.2 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
| chrX_+_105937068 | 1.56 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr7_-_41742697 | 1.11 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr12_-_52828147 | 1.05 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr3_-_172241250 | 0.89 |
ENST00000420541.2 ENST00000241261.2 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
| chr22_+_32455111 | 0.84 |
ENST00000543737.1 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr2_-_216878305 | 0.82 |
ENST00000263268.6 |
MREG |
melanoregulin |
| chr10_-_129691195 | 0.80 |
ENST00000368671.3 |
CLRN3 |
clarin 3 |
| chr10_+_24755416 | 0.77 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chr5_+_140261703 | 0.77 |
ENST00000409494.1 ENST00000289272.2 |
PCDHA13 |
protocadherin alpha 13 |
| chr7_-_16872932 | 0.77 |
ENST00000419572.2 ENST00000412973.1 |
AGR2 |
anterior gradient 2 |
| chr7_-_143105941 | 0.73 |
ENST00000275815.3 |
EPHA1 |
EPH receptor A1 |
| chr18_-_52989217 | 0.70 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr8_+_101170563 | 0.65 |
ENST00000520508.1 ENST00000388798.2 |
SPAG1 |
sperm associated antigen 1 |
| chr17_-_39646116 | 0.57 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr4_-_72649763 | 0.56 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr8_+_145215928 | 0.53 |
ENST00000528919.1 |
MROH1 |
maestro heat-like repeat family member 1 |
| chr6_+_47666275 | 0.52 |
ENST00000327753.3 ENST00000283303.2 |
GPR115 |
G protein-coupled receptor 115 |
| chr1_+_24645865 | 0.47 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_24645807 | 0.47 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr4_+_129349188 | 0.42 |
ENST00000511497.1 |
RP11-420A23.1 |
RP11-420A23.1 |
| chr1_+_46152886 | 0.41 |
ENST00000372025.4 |
TMEM69 |
transmembrane protein 69 |
| chr13_-_103719196 | 0.38 |
ENST00000245312.3 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr11_+_63606373 | 0.37 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chrX_+_65384182 | 0.35 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr15_+_35270552 | 0.35 |
ENST00000391457.2 |
AC114546.1 |
HCG37415; PRO1914; Uncharacterized protein |
| chr12_-_18243119 | 0.35 |
ENST00000538724.1 ENST00000229002.2 |
RERGL |
RERG/RAS-like |
| chr8_-_20040638 | 0.35 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chrX_+_65384052 | 0.35 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chr8_-_20040601 | 0.34 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr12_+_45686457 | 0.34 |
ENST00000441606.2 |
ANO6 |
anoctamin 6 |
| chr7_+_128784712 | 0.34 |
ENST00000289407.4 |
TSPAN33 |
tetraspanin 33 |
| chr18_-_52989525 | 0.33 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr11_+_60048004 | 0.32 |
ENST00000532114.1 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr5_-_43515231 | 0.32 |
ENST00000306862.2 |
C5orf34 |
chromosome 5 open reading frame 34 |
| chr1_-_54355430 | 0.31 |
ENST00000371399.1 ENST00000072644.1 ENST00000412288.1 |
YIPF1 |
Yip1 domain family, member 1 |
| chr7_-_130080977 | 0.29 |
ENST00000223208.5 |
CEP41 |
centrosomal protein 41kDa |
| chr4_-_76957214 | 0.28 |
ENST00000306621.3 |
CXCL11 |
chemokine (C-X-C motif) ligand 11 |
| chr20_-_18810797 | 0.28 |
ENST00000278779.4 |
C20orf78 |
chromosome 20 open reading frame 78 |
| chr1_-_106161540 | 0.28 |
ENST00000420901.1 ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1 |
RP11-251P6.1 |
| chr4_-_76944621 | 0.28 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr15_-_41836441 | 0.28 |
ENST00000567866.1 ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1 |
RNA polymerase II associated protein 1 |
| chr17_+_61086917 | 0.27 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr4_+_155484155 | 0.27 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr9_-_133814455 | 0.26 |
ENST00000448616.1 |
FIBCD1 |
fibrinogen C domain containing 1 |
| chr16_-_25122785 | 0.26 |
ENST00000563962.1 ENST00000569920.1 |
RP11-449H11.1 |
RP11-449H11.1 |
| chr5_+_121465207 | 0.24 |
ENST00000296600.4 |
ZNF474 |
zinc finger protein 474 |
| chr8_-_80993010 | 0.24 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chr2_+_27440229 | 0.24 |
ENST00000264705.4 ENST00000403525.1 |
CAD |
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr7_+_35756092 | 0.23 |
ENST00000458087.3 |
AC018647.3 |
AC018647.3 |
| chr11_+_60048053 | 0.23 |
ENST00000337908.4 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr8_+_124084899 | 0.23 |
ENST00000287380.1 ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31 |
TBC1 domain family, member 31 |
| chr11_+_120039685 | 0.23 |
ENST00000530303.1 ENST00000319763.1 |
AP000679.2 |
Uncharacterized protein |
| chr16_-_75498553 | 0.23 |
ENST00000569276.1 ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A RP11-77K12.1 |
transmembrane protein 170A Uncharacterized protein |
| chr11_+_112047087 | 0.22 |
ENST00000526088.1 ENST00000532593.1 ENST00000531169.1 |
BCO2 |
beta-carotene oxygenase 2 |
| chr7_+_35756186 | 0.22 |
ENST00000430518.1 |
AC018647.3 |
AC018647.3 |
| chr8_-_81083890 | 0.22 |
ENST00000518937.1 |
TPD52 |
tumor protein D52 |
| chr12_-_10962767 | 0.22 |
ENST00000240691.2 |
TAS2R9 |
taste receptor, type 2, member 9 |
| chr3_+_125687987 | 0.22 |
ENST00000514116.1 ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr3_+_143690640 | 0.22 |
ENST00000315691.3 |
C3orf58 |
chromosome 3 open reading frame 58 |
| chr17_+_7211656 | 0.22 |
ENST00000416016.2 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr2_+_32390925 | 0.21 |
ENST00000440718.1 ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6 |
solute carrier family 30 (zinc transporter), member 6 |
| chr6_-_133035185 | 0.21 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr10_+_76871229 | 0.21 |
ENST00000372690.3 |
SAMD8 |
sterile alpha motif domain containing 8 |
| chr12_+_1800179 | 0.20 |
ENST00000357103.4 |
ADIPOR2 |
adiponectin receptor 2 |
| chr12_-_21927736 | 0.19 |
ENST00000240662.2 |
KCNJ8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr4_+_187148556 | 0.19 |
ENST00000264690.6 ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1 |
kallikrein B, plasma (Fletcher factor) 1 |
| chr10_+_76871353 | 0.19 |
ENST00000542569.1 |
SAMD8 |
sterile alpha motif domain containing 8 |
| chr1_-_85100703 | 0.18 |
ENST00000370624.1 |
C1orf180 |
chromosome 1 open reading frame 180 |
| chr14_-_72458326 | 0.18 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr1_+_210001309 | 0.18 |
ENST00000491415.2 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
| chr6_+_125304502 | 0.18 |
ENST00000519799.1 ENST00000368414.2 ENST00000359704.2 |
RNF217 |
ring finger protein 217 |
| chr3_-_196911002 | 0.18 |
ENST00000452595.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr4_+_95972822 | 0.18 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr2_-_51259229 | 0.18 |
ENST00000405472.3 |
NRXN1 |
neurexin 1 |
| chr1_+_32379174 | 0.18 |
ENST00000391369.1 |
AL136115.1 |
HCG2032337; PRO1848; Uncharacterized protein |
| chr2_-_51259292 | 0.18 |
ENST00000401669.2 |
NRXN1 |
neurexin 1 |
| chr1_-_43919346 | 0.17 |
ENST00000372430.3 ENST00000372433.1 ENST00000372434.1 ENST00000486909.1 |
HYI |
hydroxypyruvate isomerase (putative) |
| chr16_+_71560154 | 0.17 |
ENST00000539698.3 |
CHST4 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr18_-_53177984 | 0.17 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr9_-_85678043 | 0.17 |
ENST00000376447.3 ENST00000340717.4 |
RASEF |
RAS and EF-hand domain containing |
| chr1_+_220267429 | 0.16 |
ENST00000366922.1 ENST00000302637.5 |
IARS2 |
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr16_+_71560023 | 0.16 |
ENST00000572450.1 |
CHST4 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr5_+_61874562 | 0.15 |
ENST00000334994.5 ENST00000409534.1 |
LRRC70 IPO11 |
leucine rich repeat containing 70 importin 11 |
| chr3_+_57875738 | 0.15 |
ENST00000417128.1 ENST00000438794.1 |
SLMAP |
sarcolemma associated protein |
| chr10_-_96829246 | 0.15 |
ENST00000371270.3 ENST00000535898.1 ENST00000539050.1 |
CYP2C8 |
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr3_+_140981456 | 0.15 |
ENST00000504264.1 |
ACPL2 |
acid phosphatase-like 2 |
| chr1_-_86848760 | 0.15 |
ENST00000460698.2 |
ODF2L |
outer dense fiber of sperm tails 2-like |
| chrX_-_30993201 | 0.15 |
ENST00000288422.2 ENST00000378932.2 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr9_-_133814527 | 0.15 |
ENST00000451466.1 |
FIBCD1 |
fibrinogen C domain containing 1 |
| chr12_+_64173583 | 0.15 |
ENST00000261234.6 |
TMEM5 |
transmembrane protein 5 |
| chr1_+_73771844 | 0.14 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr4_-_169239921 | 0.14 |
ENST00000514995.1 ENST00000393743.3 |
DDX60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr20_+_42984330 | 0.14 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr14_+_56584414 | 0.14 |
ENST00000559044.1 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr8_-_81083731 | 0.14 |
ENST00000379096.5 |
TPD52 |
tumor protein D52 |
| chr17_-_38821373 | 0.14 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr7_-_25268104 | 0.14 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr7_-_107204918 | 0.13 |
ENST00000297135.3 |
COG5 |
component of oligomeric golgi complex 5 |
| chr19_+_4402659 | 0.13 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr17_+_68071389 | 0.12 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_-_38956205 | 0.12 |
ENST00000306658.7 |
KRT28 |
keratin 28 |
| chrX_-_15619076 | 0.12 |
ENST00000252519.3 |
ACE2 |
angiotensin I converting enzyme 2 |
| chr13_+_25338290 | 0.12 |
ENST00000255324.5 ENST00000381921.1 ENST00000255325.6 |
RNF17 |
ring finger protein 17 |
| chr6_+_25754927 | 0.12 |
ENST00000377905.4 ENST00000439485.2 |
SLC17A4 |
solute carrier family 17, member 4 |
| chr18_+_21032781 | 0.12 |
ENST00000339486.3 |
RIOK3 |
RIO kinase 3 |
| chr2_-_32390801 | 0.12 |
ENST00000608489.1 |
RP11-563N4.1 |
RP11-563N4.1 |
| chr20_+_4702548 | 0.11 |
ENST00000305817.2 |
PRND |
prion protein 2 (dublet) |
| chr1_-_111061797 | 0.11 |
ENST00000369771.2 |
KCNA10 |
potassium voltage-gated channel, shaker-related subfamily, member 10 |
| chr18_+_3466248 | 0.11 |
ENST00000581029.1 ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4 |
RP11-838N2.4 |
| chr4_-_39979576 | 0.11 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr15_+_22736246 | 0.10 |
ENST00000316397.3 |
GOLGA6L1 |
golgin A6 family-like 1 |
| chr3_-_123710893 | 0.10 |
ENST00000467907.1 ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1 |
rhophilin associated tail protein 1 |
| chr9_-_95640218 | 0.10 |
ENST00000395506.3 ENST00000375495.3 ENST00000332591.6 |
ZNF484 |
zinc finger protein 484 |
| chr1_-_23886285 | 0.10 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr16_-_20338748 | 0.10 |
ENST00000575582.1 ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2 |
glycoprotein 2 (zymogen granule membrane) |
| chr1_+_248201474 | 0.10 |
ENST00000366479.2 |
OR2L2 |
olfactory receptor, family 2, subfamily L, member 2 |
| chr3_+_57875711 | 0.10 |
ENST00000442599.2 |
SLMAP |
sarcolemma associated protein |
| chr10_+_75504105 | 0.10 |
ENST00000535742.1 ENST00000546025.1 ENST00000345254.4 ENST00000540668.1 ENST00000339365.2 ENST00000411652.2 |
SEC24C |
SEC24 family member C |
| chr16_+_68119764 | 0.10 |
ENST00000570212.1 ENST00000562926.1 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr6_-_127840336 | 0.10 |
ENST00000525778.1 |
SOGA3 |
SOGA family member 3 |
| chr8_-_143859197 | 0.09 |
ENST00000395192.2 |
LYNX1 |
Ly6/neurotoxin 1 |
| chr10_-_24770632 | 0.09 |
ENST00000596413.1 |
AL353583.1 |
AL353583.1 |
| chr19_-_43032532 | 0.09 |
ENST00000403461.1 ENST00000352591.5 ENST00000358394.3 ENST00000403444.3 ENST00000308072.4 ENST00000599389.1 ENST00000351134.3 ENST00000161559.6 |
CEACAM1 |
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
| chr15_+_22736484 | 0.09 |
ENST00000560659.2 |
GOLGA6L1 |
golgin A6 family-like 1 |
| chr5_+_86564739 | 0.09 |
ENST00000456692.2 ENST00000512763.1 ENST00000506290.1 |
RASA1 |
RAS p21 protein activator (GTPase activating protein) 1 |
| chr21_-_27462351 | 0.09 |
ENST00000448850.1 |
APP |
amyloid beta (A4) precursor protein |
| chr16_-_20339123 | 0.09 |
ENST00000381360.5 |
GP2 |
glycoprotein 2 (zymogen granule membrane) |
| chr19_-_56343353 | 0.09 |
ENST00000592953.1 ENST00000589093.1 |
NLRP11 |
NLR family, pyrin domain containing 11 |
| chrX_+_117957741 | 0.09 |
ENST00000310164.2 |
ZCCHC12 |
zinc finger, CCHC domain containing 12 |
| chr2_-_217236750 | 0.09 |
ENST00000273067.4 |
MARCH4 |
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
| chr3_+_149191723 | 0.09 |
ENST00000305354.4 |
TM4SF4 |
transmembrane 4 L six family member 4 |
| chr11_+_46366918 | 0.09 |
ENST00000528615.1 ENST00000395574.3 |
DGKZ |
diacylglycerol kinase, zeta |
| chr4_+_155484103 | 0.08 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr4_-_164534657 | 0.08 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chrM_+_10053 | 0.08 |
ENST00000361227.2 |
MT-ND3 |
mitochondrially encoded NADH dehydrogenase 3 |
| chr6_-_27775694 | 0.08 |
ENST00000377401.2 |
HIST1H2BL |
histone cluster 1, H2bl |
| chr4_-_123377880 | 0.08 |
ENST00000226730.4 |
IL2 |
interleukin 2 |
| chr4_+_71263599 | 0.08 |
ENST00000399575.2 |
PROL1 |
proline rich, lacrimal 1 |
| chr2_-_8977714 | 0.08 |
ENST00000319688.5 ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220 |
kinase D-interacting substrate, 220kDa |
| chr13_-_45775162 | 0.08 |
ENST00000405872.1 |
KCTD4 |
potassium channel tetramerization domain containing 4 |
| chr9_-_95055956 | 0.08 |
ENST00000375629.3 ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS |
isoleucyl-tRNA synthetase |
| chr21_-_27423339 | 0.08 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr12_-_57146095 | 0.08 |
ENST00000550770.1 ENST00000338193.6 |
PRIM1 |
primase, DNA, polypeptide 1 (49kDa) |
| chrX_+_64708615 | 0.07 |
ENST00000338957.4 ENST00000423889.3 |
ZC3H12B |
zinc finger CCCH-type containing 12B |
| chr19_+_21264980 | 0.07 |
ENST00000596053.1 ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714 |
zinc finger protein 714 |
| chr6_-_166582107 | 0.07 |
ENST00000296946.2 ENST00000461348.2 ENST00000366871.3 |
T |
T, brachyury homolog (mouse) |
| chr3_+_107602030 | 0.07 |
ENST00000494231.1 |
LINC00636 |
long intergenic non-protein coding RNA 636 |
| chr9_+_134065506 | 0.07 |
ENST00000483497.2 |
NUP214 |
nucleoporin 214kDa |
| chr1_+_178694300 | 0.07 |
ENST00000367635.3 |
RALGPS2 |
Ral GEF with PH domain and SH3 binding motif 2 |
| chr14_+_73706308 | 0.07 |
ENST00000554301.1 ENST00000555445.1 |
PAPLN |
papilin, proteoglycan-like sulfated glycoprotein |
| chr1_+_158979792 | 0.07 |
ENST00000359709.3 ENST00000430894.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr11_-_60674037 | 0.07 |
ENST00000541371.1 ENST00000227524.4 |
PRPF19 |
pre-mRNA processing factor 19 |
| chr3_-_108672742 | 0.07 |
ENST00000261047.3 |
GUCA1C |
guanylate cyclase activator 1C |
| chr1_+_89246647 | 0.07 |
ENST00000544045.1 |
PKN2 |
protein kinase N2 |
| chr3_-_53925863 | 0.07 |
ENST00000541726.1 ENST00000495461.1 |
SELK |
Selenoprotein K |
| chr3_-_108672609 | 0.06 |
ENST00000393963.3 ENST00000471108.1 |
GUCA1C |
guanylate cyclase activator 1C |
| chr14_-_57272366 | 0.06 |
ENST00000554788.1 ENST00000554845.1 ENST00000408990.3 |
OTX2 |
orthodenticle homeobox 2 |
| chr9_-_95056010 | 0.06 |
ENST00000443024.2 |
IARS |
isoleucyl-tRNA synthetase |
| chr3_-_155011483 | 0.06 |
ENST00000489090.1 |
RP11-451G4.2 |
RP11-451G4.2 |
| chr10_-_103599591 | 0.06 |
ENST00000348850.5 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr15_+_40886199 | 0.06 |
ENST00000346991.5 ENST00000528975.1 ENST00000527044.1 |
CASC5 |
cancer susceptibility candidate 5 |
| chr5_-_16738451 | 0.06 |
ENST00000274203.9 ENST00000515803.1 |
MYO10 |
myosin X |
| chr7_-_111032971 | 0.06 |
ENST00000450877.1 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr16_-_21663950 | 0.06 |
ENST00000268389.4 |
IGSF6 |
immunoglobulin superfamily, member 6 |
| chr2_+_219081817 | 0.06 |
ENST00000315717.5 ENST00000420104.1 ENST00000295685.10 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr8_+_107738240 | 0.05 |
ENST00000449762.2 ENST00000297447.6 |
OXR1 |
oxidation resistance 1 |
| chr6_-_127840453 | 0.05 |
ENST00000556132.1 |
SOGA3 |
SOGA family member 3 |
| chr9_-_86571628 | 0.05 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr11_-_3400442 | 0.05 |
ENST00000429541.2 ENST00000532539.1 |
ZNF195 |
zinc finger protein 195 |
| chr2_+_38177575 | 0.05 |
ENST00000407257.1 ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2 |
regulator of microtubule dynamics 2 |
| chr4_-_69536346 | 0.05 |
ENST00000338206.5 |
UGT2B15 |
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr2_+_120687335 | 0.05 |
ENST00000544261.1 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr8_+_128426535 | 0.05 |
ENST00000465342.2 |
POU5F1B |
POU class 5 homeobox 1B |
| chr9_+_44868935 | 0.05 |
ENST00000448436.2 |
RP11-160N1.10 |
RP11-160N1.10 |
| chr17_-_72772462 | 0.05 |
ENST00000582870.1 ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9 |
N-acetyltransferase 9 (GCN5-related, putative) |
| chr19_+_21579958 | 0.04 |
ENST00000339914.6 ENST00000599461.1 |
ZNF493 |
zinc finger protein 493 |
| chr11_+_58390132 | 0.04 |
ENST00000361987.4 |
CNTF |
ciliary neurotrophic factor |
| chr11_-_3400330 | 0.04 |
ENST00000427810.2 ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195 |
zinc finger protein 195 |
| chrX_+_144908928 | 0.04 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr3_-_38992052 | 0.04 |
ENST00000302328.3 ENST00000450244.1 ENST00000444237.2 |
SCN11A |
sodium channel, voltage-gated, type XI, alpha subunit |
| chr19_+_54619125 | 0.04 |
ENST00000445811.1 ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31 |
pre-mRNA processing factor 31 |
| chr4_+_88754069 | 0.04 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr10_+_85933494 | 0.04 |
ENST00000372126.3 |
C10orf99 |
chromosome 10 open reading frame 99 |
| chr4_+_71248795 | 0.03 |
ENST00000304915.3 |
SMR3B |
submaxillary gland androgen regulated protein 3B |
| chr7_+_129984630 | 0.03 |
ENST00000355388.3 ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5 |
carboxypeptidase A5 |
| chr3_+_15045419 | 0.03 |
ENST00000406272.2 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr5_-_36001108 | 0.03 |
ENST00000333811.4 |
UGT3A1 |
UDP glycosyltransferase 3 family, polypeptide A1 |
| chr7_-_26578407 | 0.03 |
ENST00000242109.3 |
KIAA0087 |
KIAA0087 |
| chr1_-_198990166 | 0.03 |
ENST00000427439.1 |
RP11-16L9.3 |
RP11-16L9.3 |
| chr6_+_134274322 | 0.03 |
ENST00000367871.1 ENST00000237264.4 |
TBPL1 |
TBP-like 1 |
| chr4_+_186317133 | 0.03 |
ENST00000507753.1 |
ANKRD37 |
ankyrin repeat domain 37 |
| chr1_+_109419596 | 0.03 |
ENST00000435987.1 ENST00000264126.3 |
GPSM2 |
G-protein signaling modulator 2 |
| chr2_-_95787738 | 0.03 |
ENST00000272418.2 |
MRPS5 |
mitochondrial ribosomal protein S5 |
| chr4_+_88754113 | 0.02 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr17_+_58018269 | 0.02 |
ENST00000591035.1 |
RP11-178C3.1 |
Uncharacterized protein |
| chr8_+_76452097 | 0.02 |
ENST00000396423.2 |
HNF4G |
hepatocyte nuclear factor 4, gamma |
| chr1_+_158979686 | 0.02 |
ENST00000368132.3 ENST00000295809.7 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr11_+_27062860 | 0.02 |
ENST00000528583.1 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_+_12524965 | 0.02 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.3 | 1.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 1.6 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.8 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.1 | 0.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.2 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 0.8 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.3 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.6 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.9 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.3 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0043318 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) insulin catabolic process(GO:1901143) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.6 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:1903764 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0014028 | mesoderm migration involved in gastrulation(GO:0007509) notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.8 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 1.7 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.7 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.7 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.7 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.2 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 1.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |