Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
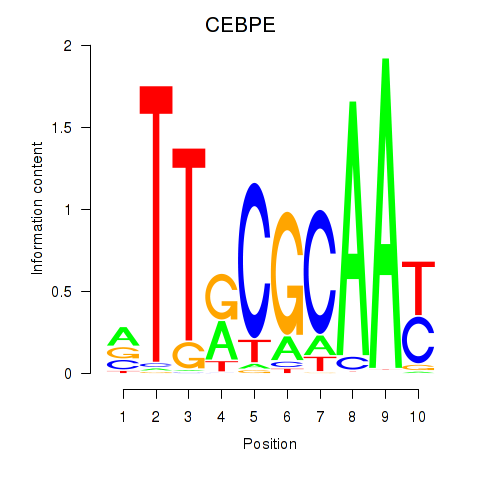
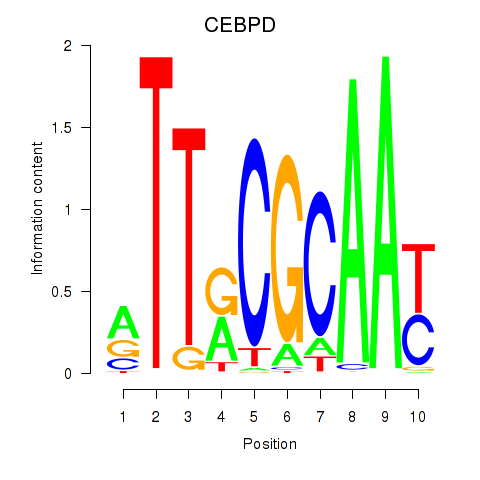
Results for CEBPE_CEBPD
Z-value: 0.75
Transcription factors associated with CEBPE_CEBPD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPE
|
ENSG00000092067.5 | CEBPE |
|
CEBPD
|
ENSG00000221869.4 | CEBPD |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPE | hg19_v2_chr14_-_23588816_23588836 | 0.51 | 1.9e-01 | Click! |
| CEBPD | hg19_v2_chr8_-_48651648_48651648 | 0.30 | 4.7e-01 | Click! |
Activity profile of CEBPE_CEBPD motif
Sorted Z-values of CEBPE_CEBPD motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPE_CEBPD
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_35992780 | 2.05 |
ENST00000593342.1 ENST00000601650.1 ENST00000408915.2 |
DMKN |
dermokine |
| chr8_+_124194875 | 2.00 |
ENST00000522648.1 ENST00000276699.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr2_-_113594279 | 1.89 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr8_+_124194752 | 1.82 |
ENST00000318462.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr11_+_18287721 | 1.73 |
ENST00000356524.4 |
SAA1 |
serum amyloid A1 |
| chr11_+_18287801 | 1.73 |
ENST00000532858.1 ENST00000405158.2 |
SAA1 |
serum amyloid A1 |
| chr11_-_18270182 | 1.62 |
ENST00000528349.1 ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2 |
serum amyloid A2 |
| chr19_-_6720686 | 1.59 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr5_+_36608422 | 1.56 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr1_-_186649543 | 1.38 |
ENST00000367468.5 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_-_123201337 | 1.23 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr20_-_43883197 | 1.16 |
ENST00000338380.2 |
SLPI |
secretory leukocyte peptidase inhibitor |
| chr12_-_123187890 | 1.14 |
ENST00000328880.5 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
| chr10_-_116444371 | 1.14 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr8_-_134309823 | 1.13 |
ENST00000414097.2 |
NDRG1 |
N-myc downstream regulated 1 |
| chr8_-_134309335 | 1.09 |
ENST00000522890.1 ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1 |
N-myc downstream regulated 1 |
| chr7_-_83824169 | 1.08 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr12_-_86230315 | 0.97 |
ENST00000361228.3 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr1_+_82266053 | 0.75 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr2_-_31637560 | 0.65 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr19_-_36004543 | 0.64 |
ENST00000339686.3 ENST00000447113.2 ENST00000440396.1 |
DMKN |
dermokine |
| chr2_-_101925055 | 0.63 |
ENST00000295317.3 |
RNF149 |
ring finger protein 149 |
| chr17_+_7348658 | 0.63 |
ENST00000570557.1 ENST00000536404.2 ENST00000576360.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chrX_+_138612889 | 0.60 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr12_+_29376673 | 0.54 |
ENST00000547116.1 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr12_+_29376592 | 0.53 |
ENST00000182377.4 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr20_+_58630972 | 0.51 |
ENST00000313426.1 |
C20orf197 |
chromosome 20 open reading frame 197 |
| chr1_+_144339738 | 0.43 |
ENST00000538264.1 |
AL592284.1 |
Protein LOC642441 |
| chr8_+_110552831 | 0.40 |
ENST00000530629.1 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr7_+_90032667 | 0.40 |
ENST00000496677.1 ENST00000287916.4 ENST00000535571.1 ENST00000394604.1 ENST00000394605.2 |
CLDN12 |
claudin 12 |
| chr4_-_139163491 | 0.39 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr20_-_22566089 | 0.38 |
ENST00000377115.4 |
FOXA2 |
forkhead box A2 |
| chr6_-_30043539 | 0.37 |
ENST00000376751.3 ENST00000244360.6 |
RNF39 |
ring finger protein 39 |
| chr10_-_113943447 | 0.36 |
ENST00000369425.1 ENST00000348367.4 ENST00000423155.1 |
GPAM |
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr19_+_56989609 | 0.36 |
ENST00000601875.1 |
ZNF667-AS1 |
ZNF667 antisense RNA 1 (head to head) |
| chr19_-_36019123 | 0.36 |
ENST00000588674.1 ENST00000452271.2 ENST00000518157.1 |
SBSN |
suprabasin |
| chr8_-_42397037 | 0.35 |
ENST00000342228.3 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
| chr20_-_52790512 | 0.34 |
ENST00000216862.3 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr7_-_121944491 | 0.34 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr6_-_99797522 | 0.34 |
ENST00000389677.5 |
FAXC |
failed axon connections homolog (Drosophila) |
| chr22_+_48972118 | 0.33 |
ENST00000358295.5 |
FAM19A5 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr5_+_49963239 | 0.32 |
ENST00000505554.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr21_+_43619796 | 0.31 |
ENST00000398457.2 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr15_+_42131011 | 0.31 |
ENST00000458483.1 |
PLA2G4B |
phospholipase A2, group IVB (cytosolic) |
| chr8_+_110552337 | 0.30 |
ENST00000337573.5 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr1_-_62784935 | 0.30 |
ENST00000354381.3 |
KANK4 |
KN motif and ankyrin repeat domains 4 |
| chr2_+_65663812 | 0.29 |
ENST00000606978.1 ENST00000377977.3 ENST00000536804.1 |
AC074391.1 |
AC074391.1 |
| chr1_-_62785054 | 0.29 |
ENST00000371153.4 |
KANK4 |
KN motif and ankyrin repeat domains 4 |
| chr2_+_113885138 | 0.29 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr1_+_196743912 | 0.28 |
ENST00000367425.4 |
CFHR3 |
complement factor H-related 3 |
| chr14_+_20937538 | 0.27 |
ENST00000361505.5 ENST00000553591.1 |
PNP |
purine nucleoside phosphorylase |
| chr1_+_196857144 | 0.26 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
| chr1_+_196743943 | 0.26 |
ENST00000471440.2 ENST00000391985.3 |
CFHR3 |
complement factor H-related 3 |
| chr2_-_46769694 | 0.24 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr3_+_130569429 | 0.24 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chrX_-_119445306 | 0.24 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chr17_+_57287228 | 0.23 |
ENST00000578922.1 ENST00000300917.5 |
SMG8 |
SMG8 nonsense mediated mRNA decay factor |
| chr16_-_84651647 | 0.23 |
ENST00000564057.1 |
COTL1 |
coactosin-like 1 (Dictyostelium) |
| chr12_+_13197218 | 0.23 |
ENST00000197268.8 |
KIAA1467 |
KIAA1467 |
| chr19_-_2042065 | 0.23 |
ENST00000591588.1 ENST00000591142.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chrX_-_119445263 | 0.22 |
ENST00000309720.5 |
TMEM255A |
transmembrane protein 255A |
| chr20_+_4667094 | 0.22 |
ENST00000424424.1 ENST00000457586.1 |
PRNP |
prion protein |
| chr3_-_158450475 | 0.22 |
ENST00000237696.5 |
RARRES1 |
retinoic acid receptor responder (tazarotene induced) 1 |
| chr11_+_112046190 | 0.21 |
ENST00000357685.5 ENST00000393032.2 ENST00000361053.4 |
BCO2 |
beta-carotene oxygenase 2 |
| chr14_-_23285069 | 0.21 |
ENST00000554758.1 ENST00000397528.4 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr16_-_84651673 | 0.21 |
ENST00000262428.4 |
COTL1 |
coactosin-like 1 (Dictyostelium) |
| chr2_-_68694390 | 0.20 |
ENST00000377957.3 |
FBXO48 |
F-box protein 48 |
| chr5_-_140013275 | 0.20 |
ENST00000512545.1 ENST00000302014.6 ENST00000401743.2 |
CD14 |
CD14 molecule |
| chr14_-_23285011 | 0.20 |
ENST00000397532.3 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_-_52521831 | 0.20 |
ENST00000371626.4 |
TXNDC12 |
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr1_+_166958346 | 0.20 |
ENST00000367872.4 |
MAEL |
maelstrom spermatogenic transposon silencer |
| chr3_-_4793274 | 0.20 |
ENST00000414938.1 |
EGOT |
eosinophil granule ontogeny transcript (non-protein coding) |
| chr1_+_166958497 | 0.19 |
ENST00000367870.2 |
MAEL |
maelstrom spermatogenic transposon silencer |
| chr14_-_81893734 | 0.19 |
ENST00000555447.1 |
STON2 |
stonin 2 |
| chr17_+_7462103 | 0.19 |
ENST00000396545.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr20_+_4666882 | 0.19 |
ENST00000379440.4 ENST00000430350.2 |
PRNP |
prion protein |
| chr1_-_111743285 | 0.18 |
ENST00000357640.4 |
DENND2D |
DENN/MADD domain containing 2D |
| chr19_+_56989485 | 0.18 |
ENST00000585445.1 ENST00000586091.1 ENST00000594783.1 ENST00000592146.1 ENST00000588158.1 ENST00000299997.4 ENST00000591797.1 |
ZNF667-AS1 |
ZNF667 antisense RNA 1 (head to head) |
| chr4_+_183164574 | 0.18 |
ENST00000511685.1 |
TENM3 |
teneurin transmembrane protein 3 |
| chr7_+_128784712 | 0.17 |
ENST00000289407.4 |
TSPAN33 |
tetraspanin 33 |
| chr2_+_183989157 | 0.17 |
ENST00000541912.1 |
NUP35 |
nucleoporin 35kDa |
| chr4_+_55095428 | 0.17 |
ENST00000508170.1 ENST00000512143.1 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr4_+_96012614 | 0.17 |
ENST00000264568.4 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr1_+_169079823 | 0.16 |
ENST00000367813.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr2_+_183989083 | 0.16 |
ENST00000295119.4 |
NUP35 |
nucleoporin 35kDa |
| chrX_+_24483338 | 0.15 |
ENST00000379162.4 ENST00000441463.2 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
| chr2_+_135676381 | 0.15 |
ENST00000537343.1 ENST00000295238.6 ENST00000264157.5 |
CCNT2 |
cyclin T2 |
| chr6_+_1389989 | 0.15 |
ENST00000259806.1 |
FOXF2 |
forkhead box F2 |
| chrX_-_73512411 | 0.15 |
ENST00000602576.1 ENST00000429124.1 |
FTX |
FTX transcript, XIST regulator (non-protein coding) |
| chr12_-_54758251 | 0.15 |
ENST00000267015.3 ENST00000551809.1 |
GPR84 |
G protein-coupled receptor 84 |
| chr3_-_10547192 | 0.15 |
ENST00000360273.2 ENST00000343816.4 |
ATP2B2 |
ATPase, Ca++ transporting, plasma membrane 2 |
| chr17_+_7461781 | 0.15 |
ENST00000349228.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr6_+_4776580 | 0.15 |
ENST00000397588.3 |
CDYL |
chromodomain protein, Y-like |
| chr16_-_70323422 | 0.14 |
ENST00000261772.8 |
AARS |
alanyl-tRNA synthetase |
| chr8_+_8559406 | 0.14 |
ENST00000519106.1 |
CLDN23 |
claudin 23 |
| chr17_+_7461849 | 0.14 |
ENST00000338784.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_+_112047087 | 0.14 |
ENST00000526088.1 ENST00000532593.1 ENST00000531169.1 |
BCO2 |
beta-carotene oxygenase 2 |
| chr8_-_102216925 | 0.14 |
ENST00000517844.1 |
ZNF706 |
zinc finger protein 706 |
| chr19_+_6887571 | 0.14 |
ENST00000250572.8 ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1 |
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr20_+_44098346 | 0.13 |
ENST00000372676.3 |
WFDC2 |
WAP four-disulfide core domain 2 |
| chrX_-_131547625 | 0.13 |
ENST00000394311.2 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr1_+_111770278 | 0.13 |
ENST00000369748.4 |
CHI3L2 |
chitinase 3-like 2 |
| chr1_+_111770232 | 0.13 |
ENST00000369744.2 |
CHI3L2 |
chitinase 3-like 2 |
| chr7_-_33080506 | 0.12 |
ENST00000381626.2 ENST00000409467.1 ENST00000449201.1 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr10_+_14880157 | 0.12 |
ENST00000378372.3 |
HSPA14 |
heat shock 70kDa protein 14 |
| chr14_+_102276192 | 0.12 |
ENST00000557714.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr11_-_77185094 | 0.12 |
ENST00000278568.4 ENST00000356341.3 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chrX_-_40506766 | 0.12 |
ENST00000378421.1 ENST00000440784.2 ENST00000327877.5 ENST00000378426.1 ENST00000378418.2 |
CXorf38 |
chromosome X open reading frame 38 |
| chr8_-_72459885 | 0.11 |
ENST00000523987.1 |
RP11-1102P16.1 |
Uncharacterized protein |
| chr9_+_130860810 | 0.11 |
ENST00000433501.1 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr1_-_151032040 | 0.11 |
ENST00000540998.1 ENST00000357235.5 |
CDC42SE1 |
CDC42 small effector 1 |
| chr9_+_130860583 | 0.11 |
ENST00000373064.5 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr20_-_5295015 | 0.11 |
ENST00000217270.3 |
PROKR2 |
prokineticin receptor 2 |
| chr20_-_43743790 | 0.11 |
ENST00000307971.4 ENST00000372789.4 |
WFDC5 |
WAP four-disulfide core domain 5 |
| chr19_+_50922187 | 0.11 |
ENST00000595883.1 ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB |
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr17_-_34417479 | 0.11 |
ENST00000225245.5 |
CCL3 |
chemokine (C-C motif) ligand 3 |
| chr4_-_155511887 | 0.11 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chr12_-_96390063 | 0.11 |
ENST00000541929.1 |
HAL |
histidine ammonia-lyase |
| chrX_-_131547596 | 0.10 |
ENST00000538204.1 ENST00000370849.3 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr14_-_72458326 | 0.10 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr4_-_2935674 | 0.10 |
ENST00000514800.1 |
MFSD10 |
major facilitator superfamily domain containing 10 |
| chr11_-_58980342 | 0.10 |
ENST00000361050.3 |
MPEG1 |
macrophage expressed 1 |
| chr11_+_82868030 | 0.10 |
ENST00000298281.4 ENST00000530660.1 |
PCF11 |
PCF11 cleavage and polyadenylation factor subunit |
| chr3_+_186435137 | 0.10 |
ENST00000447445.1 |
KNG1 |
kininogen 1 |
| chr1_-_67519782 | 0.10 |
ENST00000235345.5 |
SLC35D1 |
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
| chr17_+_72426891 | 0.10 |
ENST00000392627.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr1_-_114430169 | 0.10 |
ENST00000393316.3 |
BCL2L15 |
BCL2-like 15 |
| chr10_+_14880287 | 0.10 |
ENST00000437161.2 |
HSPA14 |
heat shock 70kDa protein 14 |
| chr17_-_41277370 | 0.09 |
ENST00000476777.1 ENST00000491747.2 ENST00000478531.1 ENST00000477152.1 ENST00000357654.3 ENST00000493795.1 ENST00000493919.1 |
BRCA1 |
breast cancer 1, early onset |
| chr15_+_45926919 | 0.09 |
ENST00000561735.1 ENST00000260324.7 |
SQRDL |
sulfide quinone reductase-like (yeast) |
| chr15_+_58430368 | 0.09 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr12_-_123215306 | 0.09 |
ENST00000356987.2 ENST00000436083.2 |
HCAR1 |
hydroxycarboxylic acid receptor 1 |
| chr3_-_58200398 | 0.09 |
ENST00000318316.3 ENST00000460422.1 ENST00000483681.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr12_+_102271129 | 0.09 |
ENST00000258534.8 |
DRAM1 |
DNA-damage regulated autophagy modulator 1 |
| chr15_+_58430567 | 0.09 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr4_+_86699834 | 0.08 |
ENST00000395183.2 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr17_-_41277467 | 0.08 |
ENST00000494123.1 ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1 |
breast cancer 1, early onset |
| chr2_+_11674213 | 0.08 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr17_+_20059302 | 0.08 |
ENST00000395530.2 |
SPECC1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr22_-_32860427 | 0.08 |
ENST00000534972.1 ENST00000397450.1 ENST00000397452.1 |
BPIFC |
BPI fold containing family C |
| chr10_-_128994422 | 0.08 |
ENST00000522781.1 |
FAM196A |
family with sequence similarity 196, member A |
| chrX_-_15620192 | 0.08 |
ENST00000427411.1 |
ACE2 |
angiotensin I converting enzyme 2 |
| chr17_-_34625719 | 0.08 |
ENST00000422211.2 ENST00000542124.1 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
| chr19_+_11350278 | 0.08 |
ENST00000252453.8 |
C19orf80 |
chromosome 19 open reading frame 80 |
| chr21_+_33671264 | 0.08 |
ENST00000339944.4 |
MRAP |
melanocortin 2 receptor accessory protein |
| chr14_-_64971288 | 0.08 |
ENST00000394715.1 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
| chr17_+_73089382 | 0.08 |
ENST00000538213.2 ENST00000584118.1 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr17_+_72427477 | 0.07 |
ENST00000342648.5 ENST00000481232.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr1_-_41950342 | 0.07 |
ENST00000372587.4 |
EDN2 |
endothelin 2 |
| chr3_-_158450231 | 0.07 |
ENST00000479756.1 |
RARRES1 |
retinoic acid receptor responder (tazarotene induced) 1 |
| chr17_-_41277317 | 0.07 |
ENST00000497488.1 ENST00000489037.1 ENST00000470026.1 ENST00000586385.1 ENST00000591534.1 ENST00000591849.1 |
BRCA1 |
breast cancer 1, early onset |
| chr8_-_103424986 | 0.07 |
ENST00000521922.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr18_+_21032781 | 0.07 |
ENST00000339486.3 |
RIOK3 |
RIO kinase 3 |
| chr9_-_86323118 | 0.07 |
ENST00000376395.4 |
UBQLN1 |
ubiquilin 1 |
| chr19_-_41388657 | 0.07 |
ENST00000301146.4 ENST00000291764.3 |
CYP2A7 |
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr11_-_104972158 | 0.07 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr1_+_92414952 | 0.07 |
ENST00000449584.1 ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT |
bromodomain, testis-specific |
| chr20_+_54987305 | 0.06 |
ENST00000371336.3 ENST00000434344.1 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr19_+_859425 | 0.06 |
ENST00000327726.6 |
CFD |
complement factor D (adipsin) |
| chr11_-_104905840 | 0.06 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr3_-_148939835 | 0.06 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr20_-_1600642 | 0.06 |
ENST00000381603.3 ENST00000381605.4 ENST00000279477.7 ENST00000568365.1 ENST00000564763.1 |
SIRPB1 RP4-576H24.4 |
signal-regulatory protein beta 1 Uncharacterized protein |
| chr2_-_112614424 | 0.06 |
ENST00000427997.1 |
ANAPC1 |
anaphase promoting complex subunit 1 |
| chr2_+_68694678 | 0.06 |
ENST00000303795.4 |
APLF |
aprataxin and PNKP like factor |
| chr16_-_67597789 | 0.06 |
ENST00000605277.1 |
CTD-2012K14.6 |
CTD-2012K14.6 |
| chr11_-_33795893 | 0.06 |
ENST00000526785.1 ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3 |
F-box protein 3 |
| chr8_-_110660999 | 0.06 |
ENST00000424158.2 ENST00000533895.1 ENST00000446070.2 ENST00000528331.1 ENST00000526302.1 ENST00000433638.1 ENST00000408908.2 ENST00000524720.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr1_-_156721502 | 0.06 |
ENST00000357325.5 |
HDGF |
hepatoma-derived growth factor |
| chr19_-_41356347 | 0.05 |
ENST00000301141.5 |
CYP2A6 |
cytochrome P450, family 2, subfamily A, polypeptide 6 |
| chr12_+_113229737 | 0.05 |
ENST00000551052.1 ENST00000415485.3 |
RPH3A |
rabphilin 3A homolog (mouse) |
| chr1_-_47697387 | 0.05 |
ENST00000371884.2 |
TAL1 |
T-cell acute lymphocytic leukemia 1 |
| chr20_+_62716348 | 0.05 |
ENST00000349451.3 |
OPRL1 |
opiate receptor-like 1 |
| chr6_-_32557610 | 0.05 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr1_-_92951607 | 0.05 |
ENST00000427103.1 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr11_-_72145426 | 0.05 |
ENST00000535990.1 ENST00000437826.2 ENST00000340729.5 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr5_+_140227357 | 0.04 |
ENST00000378122.3 |
PCDHA9 |
protocadherin alpha 9 |
| chr15_+_39873268 | 0.04 |
ENST00000397591.2 ENST00000260356.5 |
THBS1 |
thrombospondin 1 |
| chr1_+_149804218 | 0.04 |
ENST00000610125.1 |
HIST2H4A |
histone cluster 2, H4a |
| chr17_-_34524157 | 0.04 |
ENST00000378354.4 ENST00000394484.1 |
CCL3L3 |
chemokine (C-C motif) ligand 3-like 3 |
| chrX_+_49126294 | 0.04 |
ENST00000466508.1 ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F |
protein phosphatase 1, regulatory subunit 3F |
| chr19_-_41945804 | 0.04 |
ENST00000221943.9 ENST00000597457.1 ENST00000589970.1 ENST00000595425.1 ENST00000438807.3 ENST00000589102.1 ENST00000592922.2 |
ATP5SL |
ATP5S-like |
| chr2_-_201936302 | 0.04 |
ENST00000453765.1 ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B |
family with sequence similarity 126, member B |
| chr2_-_178128250 | 0.04 |
ENST00000448782.1 ENST00000446151.2 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr1_-_155829191 | 0.04 |
ENST00000437809.1 |
GON4L |
gon-4-like (C. elegans) |
| chr10_+_86004802 | 0.04 |
ENST00000359452.4 ENST00000358110.5 ENST00000372092.3 |
RGR |
retinal G protein coupled receptor |
| chr20_+_54987168 | 0.04 |
ENST00000360314.3 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr1_-_149832704 | 0.04 |
ENST00000392933.1 ENST00000369157.2 ENST00000392932.4 |
HIST2H4B |
histone cluster 2, H4b |
| chr14_-_74226961 | 0.04 |
ENST00000286523.5 ENST00000435371.1 |
ELMSAN1 |
ELM2 and Myb/SANT-like domain containing 1 |
| chr3_+_186383741 | 0.04 |
ENST00000232003.4 |
HRG |
histidine-rich glycoprotein |
| chr1_-_38100491 | 0.03 |
ENST00000356545.2 |
RSPO1 |
R-spondin 1 |
| chr2_-_178128528 | 0.03 |
ENST00000397063.4 ENST00000421929.1 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr4_-_186732048 | 0.03 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr12_+_66582919 | 0.03 |
ENST00000545837.1 ENST00000457197.2 |
IRAK3 |
interleukin-1 receptor-associated kinase 3 |
| chr1_+_209941942 | 0.03 |
ENST00000487271.1 ENST00000477431.1 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr14_-_70263979 | 0.03 |
ENST00000216540.4 |
SLC10A1 |
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr15_+_81293254 | 0.03 |
ENST00000267984.2 |
MESDC1 |
mesoderm development candidate 1 |
| chr18_-_76738843 | 0.03 |
ENST00000575722.1 ENST00000583511.1 |
RP11-849I19.1 |
RP11-849I19.1 |
| chr9_+_95726243 | 0.03 |
ENST00000416701.2 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
| chr12_-_8693469 | 0.03 |
ENST00000545274.1 ENST00000446457.2 |
CLEC4E |
C-type lectin domain family 4, member E |
| chr22_+_40297079 | 0.03 |
ENST00000344138.4 ENST00000543252.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.5 | 1.6 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.5 | 1.4 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 | 2.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 1.6 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 1.1 | GO:0048880 | sensory system development(GO:0048880) |
| 0.2 | 2.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 1.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 0.6 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.3 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.1 | 0.4 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.4 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 0.3 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.4 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.1 | 3.7 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 0.2 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.1 | 0.3 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.2 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.3 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.0 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 1.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 1.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 3.8 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 1.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030849 | autosome(GO:0030849) |
| 0.1 | 3.1 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.2 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 1.0 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 1.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 3.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 1.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 5.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 1.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.4 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 2.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.6 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |