Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
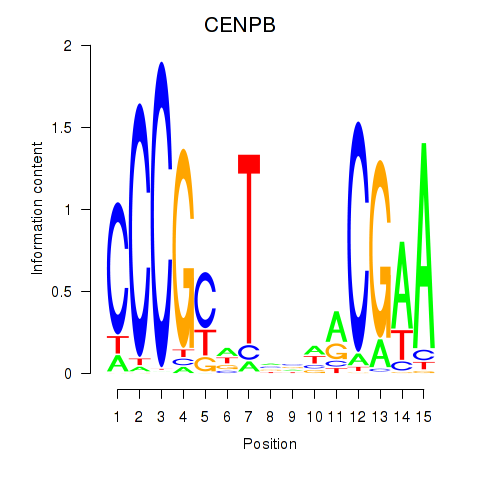
Results for CENPB
Z-value: 0.28
Transcription factors associated with CENPB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CENPB
|
ENSG00000125817.7 | CENPB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CENPB | hg19_v2_chr20_-_3767324_3767443 | 0.56 | 1.5e-01 | Click! |
Activity profile of CENPB motif
Sorted Z-values of CENPB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CENPB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_95961578 | 0.13 |
ENST00000448464.2 ENST00000342697.4 |
TP53INP1 |
tumor protein p53 inducible nuclear protein 1 |
| chr20_-_45061695 | 0.13 |
ENST00000445496.2 |
ELMO2 |
engulfment and cell motility 2 |
| chr11_+_126153001 | 0.12 |
ENST00000392678.3 ENST00000392680.2 |
TIRAP |
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chr10_+_70320413 | 0.11 |
ENST00000373644.4 |
TET1 |
tet methylcytosine dioxygenase 1 |
| chr15_-_82338460 | 0.11 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr11_+_19798964 | 0.11 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr11_+_71791849 | 0.10 |
ENST00000423494.2 ENST00000539587.1 ENST00000538478.1 ENST00000324866.7 ENST00000439209.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr6_-_57087042 | 0.09 |
ENST00000317483.3 |
RAB23 |
RAB23, member RAS oncogene family |
| chr6_+_30524663 | 0.09 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr9_-_123342415 | 0.09 |
ENST00000349780.4 ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr8_+_27168988 | 0.09 |
ENST00000397501.1 ENST00000338238.4 ENST00000544172.1 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr12_+_25348186 | 0.09 |
ENST00000555711.1 ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5 |
LYR motif containing 5 |
| chr1_+_64239657 | 0.09 |
ENST00000371080.1 ENST00000371079.1 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
| chr11_+_71791693 | 0.08 |
ENST00000289488.2 ENST00000447974.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr11_+_71791803 | 0.08 |
ENST00000539271.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr12_+_25348139 | 0.08 |
ENST00000557540.2 ENST00000381356.4 |
LYRM5 |
LYR motif containing 5 |
| chr11_+_126152954 | 0.08 |
ENST00000392679.1 |
TIRAP |
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chr6_+_30525051 | 0.08 |
ENST00000376557.3 |
PRR3 |
proline rich 3 |
| chr3_-_87040233 | 0.08 |
ENST00000398399.2 |
VGLL3 |
vestigial like 3 (Drosophila) |
| chrX_+_135230712 | 0.08 |
ENST00000535737.1 |
FHL1 |
four and a half LIM domains 1 |
| chr19_+_35168567 | 0.08 |
ENST00000457781.2 ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302 |
zinc finger protein 302 |
| chr7_+_76054224 | 0.08 |
ENST00000394857.3 |
ZP3 |
zona pellucida glycoprotein 3 (sperm receptor) |
| chr15_-_48470544 | 0.08 |
ENST00000267836.6 |
MYEF2 |
myelin expression factor 2 |
| chr11_+_71791359 | 0.07 |
ENST00000419228.1 ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr13_+_24153488 | 0.07 |
ENST00000382258.4 ENST00000382263.3 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr10_+_90639491 | 0.07 |
ENST00000371926.3 ENST00000371927.3 |
STAMBPL1 |
STAM binding protein-like 1 |
| chr15_+_45879321 | 0.07 |
ENST00000220531.3 ENST00000567461.1 |
BLOC1S6 |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr19_+_44529479 | 0.07 |
ENST00000587846.1 ENST00000187879.8 ENST00000391960.3 |
ZNF222 |
zinc finger protein 222 |
| chr11_-_560703 | 0.07 |
ENST00000441853.1 ENST00000329451.3 |
C11orf35 |
chromosome 11 open reading frame 35 |
| chr17_-_72869086 | 0.07 |
ENST00000581530.1 ENST00000420580.2 ENST00000455107.2 ENST00000413947.2 ENST00000581219.1 ENST00000582944.1 |
FDXR |
ferredoxin reductase |
| chr17_-_6947225 | 0.07 |
ENST00000574600.1 ENST00000308009.1 ENST00000447225.1 |
SLC16A11 |
solute carrier family 16, member 11 |
| chr13_+_24144509 | 0.07 |
ENST00000248484.4 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr1_-_167905225 | 0.06 |
ENST00000367846.4 |
MPC2 |
mitochondrial pyruvate carrier 2 |
| chr4_+_26321284 | 0.06 |
ENST00000506956.1 ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_+_50779071 | 0.06 |
ENST00000426751.2 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr2_+_85646054 | 0.06 |
ENST00000389938.2 |
SH2D6 |
SH2 domain containing 6 |
| chr12_-_40499661 | 0.06 |
ENST00000280871.4 |
SLC2A13 |
solute carrier family 2 (facilitated glucose transporter), member 13 |
| chr12_-_49351228 | 0.06 |
ENST00000541959.1 ENST00000447318.2 |
ARF3 |
ADP-ribosylation factor 3 |
| chr17_-_72869140 | 0.05 |
ENST00000583917.1 ENST00000293195.5 ENST00000442102.2 |
FDXR |
ferredoxin reductase |
| chr4_-_82136114 | 0.05 |
ENST00000395578.1 ENST00000418486.2 |
PRKG2 |
protein kinase, cGMP-dependent, type II |
| chr13_+_24144796 | 0.05 |
ENST00000403372.2 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr1_+_212965170 | 0.05 |
ENST00000532324.1 ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3 |
TatD DNase domain containing 3 |
| chr17_-_7531121 | 0.05 |
ENST00000573566.1 ENST00000269298.5 |
SAT2 |
spermidine/spermine N1-acetyltransferase family member 2 |
| chr15_-_48470558 | 0.05 |
ENST00000324324.7 |
MYEF2 |
myelin expression factor 2 |
| chr12_-_49351148 | 0.05 |
ENST00000398092.4 ENST00000539611.1 |
RP11-302B13.5 ARF3 |
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr2_-_241500447 | 0.05 |
ENST00000536462.1 ENST00000405002.1 ENST00000441168.1 ENST00000403283.1 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
| chr16_-_11730213 | 0.05 |
ENST00000576334.1 ENST00000574848.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr11_+_61197508 | 0.05 |
ENST00000541135.1 ENST00000301761.2 |
RP11-286N22.8 SDHAF2 |
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr4_-_170679024 | 0.04 |
ENST00000393381.2 |
C4orf27 |
chromosome 4 open reading frame 27 |
| chr15_+_63569731 | 0.04 |
ENST00000261879.5 |
APH1B |
APH1B gamma secretase subunit |
| chr8_-_145980968 | 0.04 |
ENST00000292562.7 |
ZNF251 |
zinc finger protein 251 |
| chr11_-_61582579 | 0.04 |
ENST00000539419.1 ENST00000545245.1 ENST00000545405.1 ENST00000542506.1 |
FADS1 |
fatty acid desaturase 1 |
| chr2_+_27805880 | 0.04 |
ENST00000379717.1 ENST00000355467.4 ENST00000556601.1 ENST00000416005.2 |
ZNF512 |
zinc finger protein 512 |
| chr11_+_61197572 | 0.04 |
ENST00000542074.1 ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2 |
succinate dehydrogenase complex assembly factor 2 |
| chr7_-_21985489 | 0.04 |
ENST00000356195.5 ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L |
cell division cycle associated 7-like |
| chr6_+_26124373 | 0.04 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chr7_-_158937633 | 0.04 |
ENST00000262178.2 |
VIPR2 |
vasoactive intestinal peptide receptor 2 |
| chr11_-_66115032 | 0.04 |
ENST00000311181.4 |
B3GNT1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr17_-_64188177 | 0.04 |
ENST00000535342.2 |
CEP112 |
centrosomal protein 112kDa |
| chr3_+_131100515 | 0.04 |
ENST00000537561.1 ENST00000359850.3 ENST00000521288.1 ENST00000502852.1 |
NUDT16 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr6_+_45389893 | 0.04 |
ENST00000371432.3 |
RUNX2 |
runt-related transcription factor 2 |
| chr4_-_170533723 | 0.04 |
ENST00000510533.1 ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1 |
NIMA-related kinase 1 |
| chr10_+_22605374 | 0.04 |
ENST00000448361.1 |
COMMD3 |
COMM domain containing 3 |
| chr7_-_102257139 | 0.04 |
ENST00000521076.1 ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4 |
RAS p21 protein activator 4 |
| chr3_+_157261116 | 0.04 |
ENST00000468043.1 ENST00000459838.1 ENST00000461040.1 ENST00000449199.2 ENST00000426338.2 |
C3orf55 |
chromosome 3 open reading frame 55 |
| chr14_+_24422795 | 0.04 |
ENST00000313250.5 ENST00000558263.1 ENST00000543741.2 ENST00000421831.1 ENST00000397073.2 ENST00000308178.8 ENST00000382761.3 ENST00000397075.3 ENST00000397074.3 ENST00000559632.1 ENST00000558581.1 |
DHRS4 |
dehydrogenase/reductase (SDR family) member 4 |
| chr6_-_30524951 | 0.04 |
ENST00000376621.3 |
GNL1 |
guanine nucleotide binding protein-like 1 |
| chr22_-_46646576 | 0.04 |
ENST00000314567.3 |
CDPF1 |
cysteine-rich, DPF motif domain containing 1 |
| chr15_+_63569785 | 0.04 |
ENST00000380343.4 ENST00000560353.1 |
APH1B |
APH1B gamma secretase subunit |
| chr19_-_45996465 | 0.04 |
ENST00000430715.2 |
RTN2 |
reticulon 2 |
| chr16_-_66968265 | 0.03 |
ENST00000567511.1 ENST00000422424.2 |
FAM96B |
family with sequence similarity 96, member B |
| chr16_-_30569801 | 0.03 |
ENST00000395091.2 |
ZNF764 |
zinc finger protein 764 |
| chr20_+_306177 | 0.03 |
ENST00000544632.1 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr19_+_41882466 | 0.03 |
ENST00000436170.2 |
TMEM91 |
transmembrane protein 91 |
| chr19_+_41882598 | 0.03 |
ENST00000447302.2 ENST00000544232.1 ENST00000542945.1 ENST00000540732.1 |
TMEM91 CTC-435M10.3 |
transmembrane protein 91 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial; Uncharacterized protein |
| chr16_-_30569584 | 0.03 |
ENST00000252797.2 ENST00000568114.1 |
ZNF764 AC002310.13 |
zinc finger protein 764 Uncharacterized protein |
| chr10_+_1102721 | 0.03 |
ENST00000263150.4 |
WDR37 |
WD repeat domain 37 |
| chr3_-_196669371 | 0.03 |
ENST00000427641.2 ENST00000321256.5 |
NCBP2 |
nuclear cap binding protein subunit 2, 20kDa |
| chr11_+_73019282 | 0.03 |
ENST00000263674.3 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr9_-_130953731 | 0.03 |
ENST00000420484.1 ENST00000372954.1 ENST00000541172.1 ENST00000357558.5 ENST00000325721.8 ENST00000372938.5 |
CIZ1 |
CDKN1A interacting zinc finger protein 1 |
| chrX_+_52926322 | 0.03 |
ENST00000430150.2 ENST00000452021.2 ENST00000412319.1 |
FAM156B |
family with sequence similarity 156, member B |
| chr19_-_41196458 | 0.03 |
ENST00000598779.1 |
NUMBL |
numb homolog (Drosophila)-like |
| chr7_-_150038704 | 0.03 |
ENST00000466675.1 ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2 |
retinoic acid receptor responder (tazarotene induced) 2 |
| chr1_-_151319710 | 0.03 |
ENST00000290524.4 ENST00000437327.1 ENST00000452513.2 ENST00000368870.2 ENST00000452671.2 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
| chr19_-_58609570 | 0.03 |
ENST00000600845.1 ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18 |
zinc finger and SCAN domain containing 18 |
| chr17_+_38296576 | 0.03 |
ENST00000264645.7 |
CASC3 |
cancer susceptibility candidate 3 |
| chr20_+_306221 | 0.03 |
ENST00000342665.2 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr16_-_66968055 | 0.03 |
ENST00000568572.1 |
FAM96B |
family with sequence similarity 96, member B |
| chr2_+_14772810 | 0.02 |
ENST00000295092.2 ENST00000331243.4 |
FAM84A |
family with sequence similarity 84, member A |
| chr1_+_8021954 | 0.02 |
ENST00000377491.1 ENST00000377488.1 |
PARK7 |
parkinson protein 7 |
| chrX_+_70586140 | 0.02 |
ENST00000276072.3 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr1_-_151319283 | 0.02 |
ENST00000392746.3 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
| chr22_+_20104947 | 0.02 |
ENST00000402752.1 |
RANBP1 |
RAN binding protein 1 |
| chr22_+_20105012 | 0.02 |
ENST00000331821.3 ENST00000411892.1 |
RANBP1 |
RAN binding protein 1 |
| chr11_-_18610246 | 0.02 |
ENST00000379387.4 ENST00000541984.1 |
UEVLD |
UEV and lactate/malate dehyrogenase domains |
| chr10_+_22610124 | 0.02 |
ENST00000376663.3 |
BMI1 |
BMI1 polycomb ring finger oncogene |
| chr19_+_12075844 | 0.02 |
ENST00000592625.1 ENST00000586494.1 ENST00000343949.5 ENST00000545530.1 ENST00000358987.3 |
ZNF763 |
zinc finger protein 763 |
| chr16_+_66968343 | 0.02 |
ENST00000417689.1 ENST00000561697.1 ENST00000317091.4 ENST00000566182.1 |
CES2 |
carboxylesterase 2 |
| chr16_-_20367584 | 0.02 |
ENST00000570689.1 |
UMOD |
uromodulin |
| chr14_-_60952739 | 0.02 |
ENST00000555476.1 ENST00000321731.3 |
C14orf39 |
chromosome 14 open reading frame 39 |
| chr19_+_57831829 | 0.02 |
ENST00000321545.4 |
ZNF543 |
zinc finger protein 543 |
| chr10_-_135187193 | 0.02 |
ENST00000368547.3 |
ECHS1 |
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr6_+_108487245 | 0.02 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr8_-_27168737 | 0.02 |
ENST00000521253.1 ENST00000305364.4 |
TRIM35 |
tripartite motif containing 35 |
| chr1_+_151372010 | 0.02 |
ENST00000290541.6 |
PSMB4 |
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr2_-_86790472 | 0.02 |
ENST00000409727.1 |
CHMP3 |
charged multivesicular body protein 3 |
| chr11_-_18610275 | 0.02 |
ENST00000543987.1 |
UEVLD |
UEV and lactate/malate dehyrogenase domains |
| chr15_+_49447947 | 0.02 |
ENST00000327171.3 ENST00000560654.1 |
GALK2 |
galactokinase 2 |
| chr3_-_142166846 | 0.02 |
ENST00000463916.1 ENST00000544157.1 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr7_-_5569588 | 0.02 |
ENST00000417101.1 |
ACTB |
actin, beta |
| chr16_+_25123041 | 0.02 |
ENST00000399069.3 ENST00000380966.4 |
LCMT1 |
leucine carboxyl methyltransferase 1 |
| chr20_+_21283941 | 0.02 |
ENST00000377191.3 ENST00000430571.2 |
XRN2 |
5'-3' exoribonuclease 2 |
| chr20_+_35918035 | 0.02 |
ENST00000373606.3 ENST00000397156.3 ENST00000397150.1 ENST00000397152.3 |
MANBAL |
mannosidase, beta A, lysosomal-like |
| chr5_+_137225158 | 0.02 |
ENST00000290431.5 |
PKD2L2 |
polycystic kidney disease 2-like 2 |
| chr5_+_80597419 | 0.02 |
ENST00000254037.2 ENST00000407610.3 ENST00000380199.5 |
ZCCHC9 |
zinc finger, CCHC domain containing 9 |
| chr2_+_11295624 | 0.01 |
ENST00000402361.1 ENST00000428481.1 |
PQLC3 |
PQ loop repeat containing 3 |
| chr9_-_139096955 | 0.01 |
ENST00000371748.5 |
LHX3 |
LIM homeobox 3 |
| chr14_+_100070869 | 0.01 |
ENST00000502101.2 |
RP11-543C4.1 |
RP11-543C4.1 |
| chr19_-_46195029 | 0.01 |
ENST00000588599.1 ENST00000585392.1 ENST00000590212.1 ENST00000587367.1 ENST00000391932.3 |
SNRPD2 |
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa |
| chrY_+_2709527 | 0.01 |
ENST00000250784.8 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chr20_+_3776371 | 0.01 |
ENST00000245960.5 |
CDC25B |
cell division cycle 25B |
| chrY_+_2709906 | 0.01 |
ENST00000430575.1 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chr3_-_15469006 | 0.01 |
ENST00000443029.1 ENST00000383790.3 ENST00000383789.5 |
METTL6 |
methyltransferase like 6 |
| chr15_-_49447771 | 0.01 |
ENST00000558843.1 ENST00000542928.1 ENST00000561248.1 |
COPS2 |
COP9 signalosome subunit 2 |
| chr5_-_59064458 | 0.01 |
ENST00000502575.1 ENST00000507116.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr19_+_12035878 | 0.01 |
ENST00000254321.5 ENST00000538752.1 ENST00000590798.1 |
ZNF700 ZNF763 ZNF763 |
zinc finger protein 700 zinc finger protein 763 Uncharacterized protein; Zinc finger protein 763 |
| chr12_+_110906169 | 0.01 |
ENST00000377673.5 |
FAM216A |
family with sequence similarity 216, member A |
| chr5_+_80597453 | 0.01 |
ENST00000438268.2 |
ZCCHC9 |
zinc finger, CCHC domain containing 9 |
| chr11_+_560956 | 0.01 |
ENST00000397582.3 ENST00000344375.4 ENST00000397583.3 |
RASSF7 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr2_+_11295498 | 0.01 |
ENST00000295083.3 ENST00000441908.2 |
PQLC3 |
PQ loop repeat containing 3 |
| chr6_-_46620522 | 0.01 |
ENST00000275016.2 |
CYP39A1 |
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr15_+_45879534 | 0.01 |
ENST00000564080.1 ENST00000562384.1 ENST00000569076.1 ENST00000566753.1 |
RP11-96O20.4 BLOC1S6 |
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr6_+_27356497 | 0.01 |
ENST00000244576.4 |
ZNF391 |
zinc finger protein 391 |
| chr14_+_50779029 | 0.01 |
ENST00000245448.6 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr8_-_33330595 | 0.01 |
ENST00000524021.1 ENST00000335589.3 |
FUT10 |
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
| chr11_-_82612727 | 0.01 |
ENST00000531128.1 ENST00000535099.1 ENST00000527444.1 |
PRCP |
prolylcarboxypeptidase (angiotensinase C) |
| chr22_+_30279144 | 0.01 |
ENST00000401950.2 ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3 |
myotubularin related protein 3 |
| chr12_-_49351303 | 0.01 |
ENST00000256682.4 |
ARF3 |
ADP-ribosylation factor 3 |
| chr19_-_48867291 | 0.01 |
ENST00000435956.3 |
TMEM143 |
transmembrane protein 143 |
| chr19_+_48867652 | 0.01 |
ENST00000344846.2 |
SYNGR4 |
synaptogyrin 4 |
| chr18_-_268019 | 0.00 |
ENST00000261600.6 |
THOC1 |
THO complex 1 |
| chr1_-_51810778 | 0.00 |
ENST00000413473.2 ENST00000401051.3 ENST00000527205.1 |
TTC39A |
tetratricopeptide repeat domain 39A |
| chrX_+_70586082 | 0.00 |
ENST00000373790.4 ENST00000449580.1 ENST00000423759.1 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr19_-_48867171 | 0.00 |
ENST00000377431.2 ENST00000436660.2 ENST00000541566.1 |
TMEM143 |
transmembrane protein 143 |
| chr5_+_137225125 | 0.00 |
ENST00000350250.4 ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2 |
polycystic kidney disease 2-like 2 |
| chr11_+_89764274 | 0.00 |
ENST00000448984.1 ENST00000432771.1 |
TRIM49C |
tripartite motif containing 49C |
| chr12_+_54348618 | 0.00 |
ENST00000243103.3 |
HOXC12 |
homeobox C12 |
| chr19_+_11959532 | 0.00 |
ENST00000455282.1 |
ZNF439 |
zinc finger protein 439 |
| chr7_-_102158157 | 0.00 |
ENST00000541662.1 ENST00000306682.6 ENST00000465829.1 |
RASA4B |
RAS p21 protein activator 4B |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032618 | interleukin-15 production(GO:0032618) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) positive regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000340) |
| 0.0 | 0.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.1 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.3 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.0 | GO:0009447 | putrescine catabolic process(GO:0009447) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |