Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
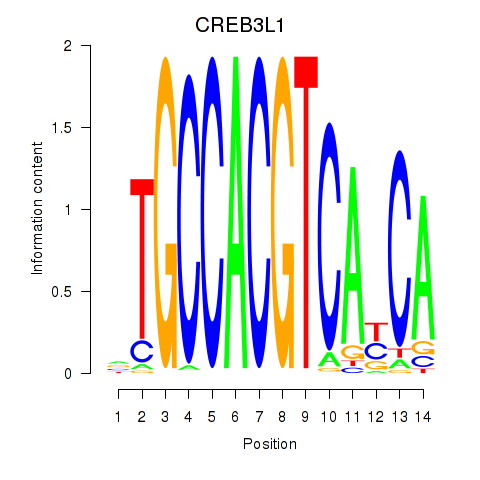
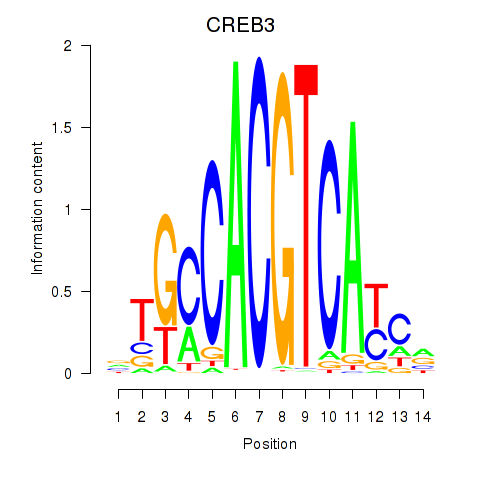
Results for CREB3L1_CREB3
Z-value: 0.39
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | CREB3L1 |
|
CREB3
|
ENSG00000107175.6 | CREB3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3 | hg19_v2_chr9_+_35732312_35732332 | 0.51 | 2.0e-01 | Click! |
| CREB3L1 | hg19_v2_chr11_+_46299199_46299233 | 0.13 | 7.7e-01 | Click! |
Activity profile of CREB3L1_CREB3 motif
Sorted Z-values of CREB3L1_CREB3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L1_CREB3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_38864041 | 0.27 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr9_+_114393581 | 0.25 |
ENST00000313525.3 |
DNAJC25 |
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr4_+_56262115 | 0.23 |
ENST00000506198.1 ENST00000381334.5 ENST00000542052.1 |
TMEM165 |
transmembrane protein 165 |
| chr3_-_156272924 | 0.22 |
ENST00000467789.1 ENST00000265044.2 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr1_+_222791417 | 0.21 |
ENST00000344922.5 ENST00000344441.6 ENST00000344507.1 |
MIA3 |
melanoma inhibitory activity family, member 3 |
| chr5_-_131132658 | 0.20 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr6_+_151561085 | 0.19 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr9_-_101984184 | 0.18 |
ENST00000476832.1 |
ALG2 |
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr19_+_50979753 | 0.17 |
ENST00000597426.1 ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10 |
ER membrane protein complex subunit 10 |
| chr15_-_28778117 | 0.17 |
ENST00000525590.2 ENST00000329523.6 |
GOLGA8G |
golgin A8 family, member G |
| chr22_+_46476192 | 0.16 |
ENST00000443490.1 |
FLJ27365 |
hsa-mir-4763 |
| chr4_+_40058411 | 0.15 |
ENST00000261435.6 ENST00000515550.1 |
N4BP2 |
NEDD4 binding protein 2 |
| chr5_+_149340282 | 0.14 |
ENST00000286298.4 |
SLC26A2 |
solute carrier family 26 (anion exchanger), member 2 |
| chr19_+_38810447 | 0.13 |
ENST00000263372.3 |
KCNK6 |
potassium channel, subfamily K, member 6 |
| chr7_+_94023873 | 0.13 |
ENST00000297268.6 |
COL1A2 |
collagen, type I, alpha 2 |
| chr7_+_6522922 | 0.13 |
ENST00000601673.1 |
FLJ20306 |
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
| chr3_-_121468602 | 0.13 |
ENST00000340645.5 |
GOLGB1 |
golgin B1 |
| chr5_-_131132614 | 0.13 |
ENST00000307968.7 ENST00000307954.8 |
FNIP1 |
folliculin interacting protein 1 |
| chr3_-_121468513 | 0.13 |
ENST00000494517.1 ENST00000393667.3 |
GOLGB1 |
golgin B1 |
| chr15_+_28623784 | 0.13 |
ENST00000526619.2 ENST00000337838.7 ENST00000532622.2 |
GOLGA8F |
golgin A8 family, member F |
| chr10_-_22292675 | 0.13 |
ENST00000376946.1 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr2_-_202645612 | 0.12 |
ENST00000409632.2 ENST00000410052.1 ENST00000467448.1 |
ALS2 |
amyotrophic lateral sclerosis 2 (juvenile) |
| chr14_-_35344093 | 0.12 |
ENST00000382422.2 |
BAZ1A |
bromodomain adjacent to zinc finger domain, 1A |
| chr11_-_3818688 | 0.12 |
ENST00000355260.3 ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98 |
nucleoporin 98kDa |
| chr3_-_194119083 | 0.12 |
ENST00000401815.1 |
GP5 |
glycoprotein V (platelet) |
| chr2_+_183580954 | 0.12 |
ENST00000264065.7 |
DNAJC10 |
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr8_-_38126675 | 0.12 |
ENST00000531823.1 ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B |
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr9_-_127703333 | 0.12 |
ENST00000373555.4 |
GOLGA1 |
golgin A1 |
| chr4_+_6202448 | 0.12 |
ENST00000508601.1 |
RP11-586D19.1 |
RP11-586D19.1 |
| chr16_-_11836595 | 0.12 |
ENST00000356957.3 ENST00000283033.5 |
TXNDC11 |
thioredoxin domain containing 11 |
| chr18_+_56806701 | 0.12 |
ENST00000587834.1 |
SEC11C |
SEC11 homolog C (S. cerevisiae) |
| chr4_-_169931231 | 0.11 |
ENST00000504561.1 |
CBR4 |
carbonyl reductase 4 |
| chrX_-_8139308 | 0.11 |
ENST00000317103.4 |
VCX2 |
variable charge, X-linked 2 |
| chr12_+_113376249 | 0.10 |
ENST00000551007.1 ENST00000548514.1 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr16_+_53164833 | 0.10 |
ENST00000564845.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr19_-_52097613 | 0.10 |
ENST00000301439.3 |
AC018755.1 |
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
| chr2_+_27255806 | 0.10 |
ENST00000238788.9 ENST00000404032.3 |
TMEM214 |
transmembrane protein 214 |
| chr20_-_17662878 | 0.10 |
ENST00000377813.1 ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1 |
ribosome binding protein 1 |
| chr5_-_131562935 | 0.10 |
ENST00000379104.2 ENST00000379100.2 ENST00000428369.1 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr7_+_150929550 | 0.09 |
ENST00000482173.1 ENST00000495645.1 ENST00000035307.2 |
CHPF2 |
chondroitin polymerizing factor 2 |
| chr11_-_67141090 | 0.09 |
ENST00000312438.7 |
CLCF1 |
cardiotrophin-like cytokine factor 1 |
| chr7_-_30066233 | 0.09 |
ENST00000222803.5 |
FKBP14 |
FK506 binding protein 14, 22 kDa |
| chr5_-_94890648 | 0.09 |
ENST00000513823.1 ENST00000514952.1 ENST00000358746.2 |
TTC37 |
tetratricopeptide repeat domain 37 |
| chr15_+_41952591 | 0.09 |
ENST00000566718.1 ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA |
MGA, MAX dimerization protein |
| chr20_-_17662705 | 0.09 |
ENST00000455029.2 |
RRBP1 |
ribosome binding protein 1 |
| chr5_-_19988339 | 0.09 |
ENST00000382275.1 |
CDH18 |
cadherin 18, type 2 |
| chr7_-_100493744 | 0.09 |
ENST00000428317.1 ENST00000441605.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr8_-_38126635 | 0.09 |
ENST00000529359.1 |
PPAPDC1B |
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr15_+_84904525 | 0.09 |
ENST00000510439.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr19_+_55476620 | 0.09 |
ENST00000543010.1 ENST00000391721.4 ENST00000339757.7 |
NLRP2 |
NLR family, pyrin domain containing 2 |
| chr12_+_49372251 | 0.09 |
ENST00000293549.3 |
WNT1 |
wingless-type MMTV integration site family, member 1 |
| chr2_-_114514181 | 0.09 |
ENST00000409342.1 |
SLC35F5 |
solute carrier family 35, member F5 |
| chr14_+_50087468 | 0.09 |
ENST00000305386.2 |
MGAT2 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr8_-_29208183 | 0.08 |
ENST00000240100.2 |
DUSP4 |
dual specificity phosphatase 4 |
| chr5_-_138210977 | 0.08 |
ENST00000274711.6 ENST00000521094.2 |
LRRTM2 |
leucine rich repeat transmembrane neuronal 2 |
| chr5_+_133984462 | 0.08 |
ENST00000398844.2 ENST00000322887.4 |
SEC24A |
SEC24 family member A |
| chr1_+_11866270 | 0.08 |
ENST00000376497.3 ENST00000376487.3 ENST00000376496.3 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr3_+_3168692 | 0.08 |
ENST00000402675.1 ENST00000413000.1 |
TRNT1 |
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr19_-_10341948 | 0.08 |
ENST00000590320.1 ENST00000592342.1 ENST00000588952.1 |
S1PR2 DNMT1 |
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr1_-_155145721 | 0.08 |
ENST00000295682.4 |
KRTCAP2 |
keratinocyte associated protein 2 |
| chr19_-_4670345 | 0.08 |
ENST00000599630.1 ENST00000262947.3 |
C19orf10 |
chromosome 19 open reading frame 10 |
| chr5_-_114961858 | 0.08 |
ENST00000282382.4 ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2 TMED7 TICAM2 |
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr7_+_99070464 | 0.08 |
ENST00000331410.5 ENST00000483089.1 ENST00000448667.1 ENST00000493485.1 |
ZNF789 |
zinc finger protein 789 |
| chr2_-_120980939 | 0.08 |
ENST00000426077.2 |
TMEM185B |
transmembrane protein 185B |
| chr19_-_58459039 | 0.08 |
ENST00000282308.3 ENST00000598928.1 |
ZNF256 |
zinc finger protein 256 |
| chr8_+_140943416 | 0.08 |
ENST00000507535.3 |
C8orf17 |
chromosome 8 open reading frame 17 |
| chr3_+_47422485 | 0.08 |
ENST00000431726.1 ENST00000456221.1 ENST00000265562.4 |
PTPN23 |
protein tyrosine phosphatase, non-receptor type 23 |
| chr5_-_131563501 | 0.08 |
ENST00000401867.1 ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr19_-_15236562 | 0.08 |
ENST00000263383.3 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chrX_-_34150428 | 0.07 |
ENST00000346193.3 |
FAM47A |
family with sequence similarity 47, member A |
| chr19_-_48894104 | 0.07 |
ENST00000597017.1 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr4_+_165675269 | 0.07 |
ENST00000507311.1 |
RP11-294O2.2 |
RP11-294O2.2 |
| chr16_+_19125252 | 0.07 |
ENST00000566735.1 ENST00000381440.3 |
ITPRIPL2 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chr2_-_43823093 | 0.07 |
ENST00000405006.4 |
THADA |
thyroid adenoma associated |
| chr10_+_27444268 | 0.07 |
ENST00000375940.4 ENST00000342386.6 |
MASTL |
microtubule associated serine/threonine kinase-like |
| chr11_+_67159416 | 0.07 |
ENST00000307980.2 ENST00000544620.1 |
RAD9A |
RAD9 homolog A (S. pombe) |
| chr1_+_92414952 | 0.07 |
ENST00000449584.1 ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT |
bromodomain, testis-specific |
| chr10_-_27444143 | 0.07 |
ENST00000477432.1 |
YME1L1 |
YME1-like 1 ATPase |
| chr6_+_132891461 | 0.07 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr3_-_156272872 | 0.07 |
ENST00000476217.1 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr14_+_24600484 | 0.07 |
ENST00000267426.5 |
FITM1 |
fat storage-inducing transmembrane protein 1 |
| chr7_+_116593953 | 0.07 |
ENST00000397750.3 |
ST7-OT4 |
ST7 overlapping transcript 4 |
| chr3_+_100211412 | 0.07 |
ENST00000323523.4 ENST00000403410.1 ENST00000449609.1 |
TMEM45A |
transmembrane protein 45A |
| chr9_+_126777676 | 0.06 |
ENST00000488674.2 |
LHX2 |
LIM homeobox 2 |
| chr12_-_106641728 | 0.06 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chr1_-_43833628 | 0.06 |
ENST00000413844.2 ENST00000372458.3 |
ELOVL1 |
ELOVL fatty acid elongase 1 |
| chr3_-_118959716 | 0.06 |
ENST00000467604.1 ENST00000491906.1 ENST00000475803.1 ENST00000479150.1 ENST00000470111.1 ENST00000459820.1 |
B4GALT4 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr12_+_2921788 | 0.06 |
ENST00000228799.2 ENST00000419778.2 ENST00000542548.1 |
ITFG2 |
integrin alpha FG-GAP repeat containing 2 |
| chr11_-_68039364 | 0.06 |
ENST00000533310.1 ENST00000304271.6 ENST00000527280.1 |
C11orf24 |
chromosome 11 open reading frame 24 |
| chr6_-_7313381 | 0.06 |
ENST00000489567.1 ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1 |
signal sequence receptor, alpha |
| chr3_-_118959733 | 0.06 |
ENST00000459778.1 ENST00000359213.3 |
B4GALT4 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr6_-_44225231 | 0.06 |
ENST00000538577.1 ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr12_+_56624436 | 0.06 |
ENST00000266980.4 ENST00000437277.1 |
SLC39A5 |
solute carrier family 39 (zinc transporter), member 5 |
| chr6_-_109702885 | 0.06 |
ENST00000504373.1 |
CD164 |
CD164 molecule, sialomucin |
| chr11_-_102576537 | 0.06 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr19_-_15236470 | 0.06 |
ENST00000533747.1 ENST00000598709.1 ENST00000534378.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr17_+_73089382 | 0.06 |
ENST00000538213.2 ENST00000584118.1 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr1_-_113498943 | 0.06 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chrX_-_46618490 | 0.06 |
ENST00000328306.4 |
SLC9A7 |
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr15_-_72668805 | 0.06 |
ENST00000268097.5 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr11_+_69455855 | 0.06 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr18_+_76829441 | 0.06 |
ENST00000458297.2 |
ATP9B |
ATPase, class II, type 9B |
| chr7_-_65958525 | 0.06 |
ENST00000450353.1 ENST00000412091.1 |
GS1-124K5.3 |
GS1-124K5.3 |
| chr13_+_111766897 | 0.06 |
ENST00000317133.5 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr4_+_141445311 | 0.06 |
ENST00000323570.3 ENST00000511887.2 |
ELMOD2 |
ELMO/CED-12 domain containing 2 |
| chr11_+_32112431 | 0.06 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr9_-_131709858 | 0.06 |
ENST00000372586.3 |
DOLK |
dolichol kinase |
| chr12_-_56123444 | 0.06 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr8_+_64081156 | 0.06 |
ENST00000517371.1 |
YTHDF3 |
YTH domain family, member 3 |
| chr5_+_109025067 | 0.06 |
ENST00000261483.4 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
| chr5_+_94890840 | 0.06 |
ENST00000504763.1 |
ARSK |
arylsulfatase family, member K |
| chr1_-_113498616 | 0.06 |
ENST00000433570.4 ENST00000538576.1 ENST00000458229.1 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr9_-_136242956 | 0.06 |
ENST00000371989.3 ENST00000485435.2 |
SURF4 |
surfeit 4 |
| chr12_+_1100449 | 0.05 |
ENST00000360905.4 |
ERC1 |
ELKS/RAB6-interacting/CAST family member 1 |
| chr15_-_72668185 | 0.05 |
ENST00000457859.2 ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr17_-_15466742 | 0.05 |
ENST00000584811.1 ENST00000419890.2 |
TVP23C |
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr20_-_5100591 | 0.05 |
ENST00000379143.5 |
PCNA |
proliferating cell nuclear antigen |
| chr4_+_75023816 | 0.05 |
ENST00000395759.2 ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr16_-_73082274 | 0.05 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr1_+_11866207 | 0.05 |
ENST00000312413.6 ENST00000346436.6 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr12_+_50355647 | 0.05 |
ENST00000293599.6 |
AQP5 |
aquaporin 5 |
| chrX_-_6453159 | 0.05 |
ENST00000381089.3 ENST00000398729.1 |
VCX3A |
variable charge, X-linked 3A |
| chr10_-_98346801 | 0.05 |
ENST00000371142.4 |
TM9SF3 |
transmembrane 9 superfamily member 3 |
| chr7_+_128379346 | 0.05 |
ENST00000535011.2 ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU |
calumenin |
| chr20_-_35402123 | 0.05 |
ENST00000373740.3 ENST00000426836.1 ENST00000373745.3 ENST00000448110.2 ENST00000438549.1 ENST00000447406.1 ENST00000373750.4 ENST00000373734.4 |
DSN1 |
DSN1, MIS12 kinetochore complex component |
| chr8_-_57906362 | 0.05 |
ENST00000262644.4 |
IMPAD1 |
inositol monophosphatase domain containing 1 |
| chr15_-_23692381 | 0.05 |
ENST00000567107.1 ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2 |
golgin A6 family-like 2 |
| chrX_-_19689106 | 0.05 |
ENST00000379716.1 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr11_-_64646086 | 0.05 |
ENST00000320631.3 |
EHD1 |
EH-domain containing 1 |
| chr12_+_1100370 | 0.05 |
ENST00000543086.3 ENST00000546231.2 ENST00000397203.2 |
ERC1 |
ELKS/RAB6-interacting/CAST family member 1 |
| chr12_-_56709674 | 0.05 |
ENST00000551286.1 ENST00000549318.1 |
CNPY2 RP11-977G19.10 |
canopy FGF signaling regulator 2 Uncharacterized protein |
| chr11_-_3818932 | 0.05 |
ENST00000324932.7 ENST00000359171.4 |
NUP98 |
nucleoporin 98kDa |
| chr1_-_241520525 | 0.05 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr19_-_41196534 | 0.05 |
ENST00000252891.4 |
NUMBL |
numb homolog (Drosophila)-like |
| chr7_-_100860851 | 0.05 |
ENST00000223127.3 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr11_+_17373273 | 0.05 |
ENST00000338965.4 |
NCR3LG1 |
natural killer cell cytotoxicity receptor 3 ligand 1 |
| chr3_+_169684553 | 0.05 |
ENST00000337002.4 ENST00000480708.1 |
SEC62 |
SEC62 homolog (S. cerevisiae) |
| chr2_-_43823119 | 0.05 |
ENST00000403856.1 ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA |
thyroid adenoma associated |
| chr2_+_192542850 | 0.05 |
ENST00000410026.2 |
NABP1 |
nucleic acid binding protein 1 |
| chr1_+_160313062 | 0.05 |
ENST00000294785.5 ENST00000368063.1 ENST00000437169.1 |
NCSTN |
nicastrin |
| chr6_+_30908747 | 0.05 |
ENST00000462446.1 ENST00000304311.2 |
DPCR1 |
diffuse panbronchiolitis critical region 1 |
| chr20_-_48782639 | 0.05 |
ENST00000435301.2 |
RP11-112L6.3 |
RP11-112L6.3 |
| chr8_-_71519889 | 0.05 |
ENST00000521425.1 |
TRAM1 |
translocation associated membrane protein 1 |
| chr12_-_49318715 | 0.05 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr4_-_83350580 | 0.05 |
ENST00000349655.4 ENST00000602300.1 |
HNRNPDL |
heterogeneous nuclear ribonucleoprotein D-like |
| chr16_+_84209539 | 0.05 |
ENST00000569735.1 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
| chr2_+_220042933 | 0.05 |
ENST00000430297.2 |
FAM134A |
family with sequence similarity 134, member A |
| chr7_+_128379449 | 0.04 |
ENST00000479257.1 |
CALU |
calumenin |
| chr16_+_699319 | 0.04 |
ENST00000549091.1 ENST00000293879.4 |
WDR90 |
WD repeat domain 90 |
| chrX_+_153060090 | 0.04 |
ENST00000370086.3 ENST00000370085.3 |
SSR4 |
signal sequence receptor, delta |
| chr7_+_102073966 | 0.04 |
ENST00000495936.1 ENST00000356387.2 ENST00000478730.2 ENST00000468241.1 ENST00000403646.3 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
| chr8_-_134115118 | 0.04 |
ENST00000395352.3 ENST00000338087.5 |
SLA |
Src-like-adaptor |
| chr8_+_64081214 | 0.04 |
ENST00000542911.2 |
YTHDF3 |
YTH domain family, member 3 |
| chr11_-_118972575 | 0.04 |
ENST00000432443.2 |
DPAGT1 |
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr22_-_36903101 | 0.04 |
ENST00000397224.4 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr12_+_56552128 | 0.04 |
ENST00000548580.1 ENST00000293422.5 ENST00000348108.4 ENST00000549017.1 ENST00000549566.1 ENST00000536128.1 ENST00000547649.1 ENST00000547408.1 ENST00000551589.1 ENST00000549392.1 ENST00000548400.1 ENST00000548293.1 |
MYL6 |
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr19_-_15236173 | 0.04 |
ENST00000527093.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr1_+_45205478 | 0.04 |
ENST00000452259.1 ENST00000372224.4 |
KIF2C |
kinesin family member 2C |
| chr13_-_52027134 | 0.04 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr5_-_140700322 | 0.04 |
ENST00000313368.5 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr5_-_54603368 | 0.04 |
ENST00000508346.1 ENST00000251636.5 |
DHX29 |
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chrX_-_118699325 | 0.04 |
ENST00000320339.4 ENST00000371594.4 ENST00000536133.1 |
CXorf56 |
chromosome X open reading frame 56 |
| chr5_-_150080472 | 0.04 |
ENST00000521464.1 ENST00000518917.1 ENST00000447771.2 ENST00000540000.1 ENST00000199814.4 |
RBM22 |
RNA binding motif protein 22 |
| chr17_-_15466850 | 0.04 |
ENST00000438826.3 ENST00000225576.3 ENST00000519970.1 ENST00000518321.1 ENST00000428082.2 ENST00000522212.2 |
TVP23C TVP23C-CDRT4 |
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) TVP23C-CDRT4 readthrough |
| chr7_+_89975979 | 0.04 |
ENST00000257659.8 ENST00000222511.6 ENST00000417207.1 |
GTPBP10 |
GTP-binding protein 10 (putative) |
| chr1_+_45205498 | 0.04 |
ENST00000372218.4 |
KIF2C |
kinesin family member 2C |
| chr11_+_3819049 | 0.04 |
ENST00000396986.2 ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chr20_+_55926583 | 0.04 |
ENST00000395840.2 |
RAE1 |
ribonucleic acid export 1 |
| chr20_+_31595406 | 0.04 |
ENST00000170150.3 |
BPIFB2 |
BPI fold containing family B, member 2 |
| chr2_-_202645835 | 0.04 |
ENST00000264276.6 |
ALS2 |
amyotrophic lateral sclerosis 2 (juvenile) |
| chr11_+_47430133 | 0.04 |
ENST00000531974.1 ENST00000531419.1 ENST00000531865.1 ENST00000362021.4 ENST00000354884.4 |
SLC39A13 |
solute carrier family 39 (zinc transporter), member 13 |
| chr1_-_160313025 | 0.04 |
ENST00000368069.3 ENST00000241704.7 |
COPA |
coatomer protein complex, subunit alpha |
| chr1_-_153940097 | 0.04 |
ENST00000413622.1 ENST00000310483.6 |
SLC39A1 |
solute carrier family 39 (zinc transporter), member 1 |
| chr20_+_33146510 | 0.04 |
ENST00000397709.1 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
| chr17_+_4046462 | 0.04 |
ENST00000577075.2 ENST00000575251.1 ENST00000301391.3 |
CYB5D2 |
cytochrome b5 domain containing 2 |
| chrX_+_47441712 | 0.03 |
ENST00000218388.4 ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr5_-_150138551 | 0.03 |
ENST00000446090.2 ENST00000447998.2 |
DCTN4 |
dynactin 4 (p62) |
| chrX_+_152240819 | 0.03 |
ENST00000421798.3 ENST00000535416.1 |
PNMA6C PNMA6A |
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr1_-_101360331 | 0.03 |
ENST00000416479.1 ENST00000370113.3 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
| chr9_+_112542572 | 0.03 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr7_-_73184588 | 0.03 |
ENST00000395145.2 |
CLDN3 |
claudin 3 |
| chr17_-_7218403 | 0.03 |
ENST00000570780.1 |
GPS2 |
G protein pathway suppressor 2 |
| chr3_-_4508925 | 0.03 |
ENST00000534863.1 ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1 |
sulfatase modifying factor 1 |
| chr6_-_101329191 | 0.03 |
ENST00000324723.6 ENST00000369162.2 ENST00000522650.1 |
ASCC3 |
activating signal cointegrator 1 complex subunit 3 |
| chr11_-_14521379 | 0.03 |
ENST00000249923.3 ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1 |
coatomer protein complex, subunit beta 1 |
| chr19_-_41196458 | 0.03 |
ENST00000598779.1 |
NUMBL |
numb homolog (Drosophila)-like |
| chr17_+_55163075 | 0.03 |
ENST00000571629.1 ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr5_-_39425290 | 0.03 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chrX_+_153059608 | 0.03 |
ENST00000370087.1 |
SSR4 |
signal sequence receptor, delta |
| chr5_-_89825328 | 0.03 |
ENST00000500869.2 ENST00000315948.6 ENST00000509384.1 |
LYSMD3 |
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr3_-_119182523 | 0.03 |
ENST00000319172.5 |
TMEM39A |
transmembrane protein 39A |
| chr17_-_7218631 | 0.03 |
ENST00000577040.2 ENST00000389167.5 ENST00000391950.3 |
GPS2 |
G protein pathway suppressor 2 |
| chr4_-_119757239 | 0.03 |
ENST00000280551.6 |
SEC24D |
SEC24 family member D |
| chr5_-_114961673 | 0.03 |
ENST00000333314.3 |
TMED7-TICAM2 |
TMED7-TICAM2 readthrough |
| chr2_-_33824382 | 0.03 |
ENST00000238823.8 |
FAM98A |
family with sequence similarity 98, member A |
| chr3_-_28390415 | 0.03 |
ENST00000414162.1 ENST00000420543.2 |
AZI2 |
5-azacytidine induced 2 |
| chr7_-_138720763 | 0.03 |
ENST00000275766.1 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
| chr11_+_125496124 | 0.03 |
ENST00000533778.2 ENST00000534070.1 |
CHEK1 |
checkpoint kinase 1 |
| chr20_-_61051026 | 0.03 |
ENST00000252997.2 |
GATA5 |
GATA binding protein 5 |
| chr2_-_33824336 | 0.03 |
ENST00000431950.1 ENST00000403368.1 ENST00000441530.2 |
FAM98A |
family with sequence similarity 98, member A |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.3 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0061183 | Spemann organizer formation(GO:0060061) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.2 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |