Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
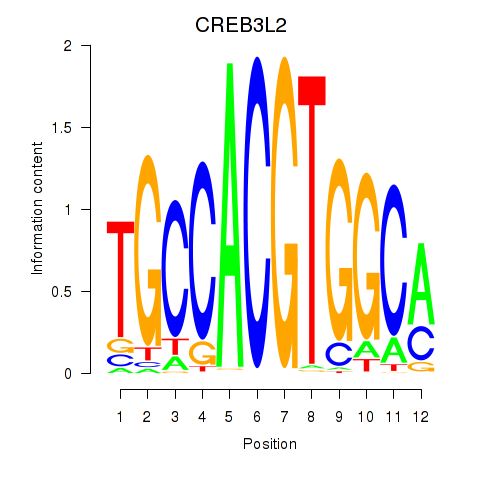
Results for CREB3L2
Z-value: 0.64
Transcription factors associated with CREB3L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L2
|
ENSG00000182158.10 | CREB3L2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L2 | hg19_v2_chr7_-_137686791_137686821 | 0.89 | 2.7e-03 | Click! |
Activity profile of CREB3L2 motif
Sorted Z-values of CREB3L2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_178789220 | 0.79 |
ENST00000414084.1 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr3_+_141144963 | 0.67 |
ENST00000510726.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr5_-_150537279 | 0.67 |
ENST00000517486.1 ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6 |
annexin A6 |
| chr12_+_50355647 | 0.62 |
ENST00000293599.6 |
AQP5 |
aquaporin 5 |
| chr7_+_29603394 | 0.59 |
ENST00000319694.2 |
PRR15 |
proline rich 15 |
| chr10_+_89419370 | 0.57 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr10_+_131265443 | 0.51 |
ENST00000306010.7 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
| chr2_+_46926048 | 0.50 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr11_+_67776012 | 0.49 |
ENST00000539229.1 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
| chr22_+_38864041 | 0.46 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr16_+_85061367 | 0.46 |
ENST00000538274.1 ENST00000258180.3 |
KIAA0513 |
KIAA0513 |
| chr2_+_217498105 | 0.45 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr2_+_46926326 | 0.44 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr9_-_113800341 | 0.44 |
ENST00000358883.4 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr9_+_92219919 | 0.41 |
ENST00000252506.6 ENST00000375769.1 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
| chr1_-_145039835 | 0.41 |
ENST00000533259.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr12_-_76953453 | 0.41 |
ENST00000549570.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr20_-_2821271 | 0.38 |
ENST00000448755.1 ENST00000360652.2 |
PCED1A |
PC-esterase domain containing 1A |
| chrX_+_55744166 | 0.35 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr19_+_49458107 | 0.35 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chr17_-_30185946 | 0.35 |
ENST00000579741.1 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
| chrX_+_55744228 | 0.33 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr10_-_104474128 | 0.33 |
ENST00000260746.5 |
ARL3 |
ADP-ribosylation factor-like 3 |
| chr17_+_39969183 | 0.32 |
ENST00000321562.4 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr1_-_241520525 | 0.32 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr7_-_8302207 | 0.30 |
ENST00000407906.1 |
ICA1 |
islet cell autoantigen 1, 69kDa |
| chr17_-_30185971 | 0.30 |
ENST00000378634.2 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
| chrX_+_153686614 | 0.30 |
ENST00000369682.3 |
PLXNA3 |
plexin A3 |
| chr11_-_57282349 | 0.29 |
ENST00000528450.1 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr6_+_32811885 | 0.29 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr1_-_145039949 | 0.28 |
ENST00000313382.9 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr14_-_74551096 | 0.28 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr22_-_36903101 | 0.27 |
ENST00000397224.4 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr19_+_1269324 | 0.27 |
ENST00000589710.1 ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP |
cold inducible RNA binding protein |
| chr11_+_6502675 | 0.27 |
ENST00000254616.6 ENST00000530751.1 |
TIMM10B |
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr6_+_151561085 | 0.27 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr6_+_32811861 | 0.27 |
ENST00000453426.1 |
TAPSAR1 |
TAP1 and PSMB8 antisense RNA 1 |
| chr22_-_36902522 | 0.26 |
ENST00000397223.4 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr16_-_67185117 | 0.26 |
ENST00000449549.3 |
B3GNT9 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr2_-_101767715 | 0.26 |
ENST00000376840.4 ENST00000409318.1 |
TBC1D8 |
TBC1 domain family, member 8 (with GRAM domain) |
| chr14_-_74551172 | 0.26 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr9_-_113800981 | 0.26 |
ENST00000538760.1 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr5_-_176730676 | 0.25 |
ENST00000393611.2 ENST00000303251.6 ENST00000303270.6 |
RAB24 |
RAB24, member RAS oncogene family |
| chr1_+_46016703 | 0.25 |
ENST00000481885.1 ENST00000351829.4 ENST00000471651.1 |
AKR1A1 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr22_-_36903069 | 0.24 |
ENST00000216187.6 ENST00000423980.1 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr5_-_114880533 | 0.24 |
ENST00000274457.3 |
FEM1C |
fem-1 homolog c (C. elegans) |
| chr19_-_12946236 | 0.24 |
ENST00000589272.1 ENST00000393233.2 |
RTBDN |
retbindin |
| chr17_-_30186328 | 0.23 |
ENST00000302362.6 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
| chr7_+_120702819 | 0.23 |
ENST00000423795.1 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr10_-_49459800 | 0.22 |
ENST00000305531.3 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr16_-_4897266 | 0.22 |
ENST00000591451.1 ENST00000436648.5 ENST00000381983.3 ENST00000588297.1 ENST00000321919.9 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr12_-_56122761 | 0.22 |
ENST00000552164.1 ENST00000420846.3 ENST00000257857.4 |
CD63 |
CD63 molecule |
| chr19_-_12946215 | 0.21 |
ENST00000591512.1 ENST00000587549.1 ENST00000322912.5 |
RTBDN |
retbindin |
| chr20_+_25228669 | 0.21 |
ENST00000216962.4 |
PYGB |
phosphorylase, glycogen; brain |
| chr6_+_127588020 | 0.21 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr17_+_19552036 | 0.20 |
ENST00000581518.1 ENST00000395575.2 ENST00000584332.2 ENST00000339618.4 ENST00000579855.1 |
ALDH3A2 |
aldehyde dehydrogenase 3 family, member A2 |
| chr7_+_101917407 | 0.20 |
ENST00000487284.1 |
CUX1 |
cut-like homeobox 1 |
| chr12_-_76953513 | 0.20 |
ENST00000547540.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr4_-_83812248 | 0.20 |
ENST00000514326.1 ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr17_+_57408994 | 0.20 |
ENST00000312655.4 |
YPEL2 |
yippee-like 2 (Drosophila) |
| chr6_+_31802685 | 0.20 |
ENST00000375639.2 ENST00000375638.3 ENST00000375635.2 ENST00000375642.2 ENST00000395789.1 |
C6orf48 |
chromosome 6 open reading frame 48 |
| chr15_+_62359175 | 0.20 |
ENST00000355522.5 |
C2CD4A |
C2 calcium-dependent domain containing 4A |
| chr22_-_24093267 | 0.20 |
ENST00000341976.3 |
ZNF70 |
zinc finger protein 70 |
| chr15_+_41913690 | 0.20 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chr4_-_83812402 | 0.19 |
ENST00000395310.2 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr15_-_40213080 | 0.19 |
ENST00000561100.1 |
GPR176 |
G protein-coupled receptor 176 |
| chr22_-_41636929 | 0.19 |
ENST00000216241.9 |
CHADL |
chondroadherin-like |
| chr5_-_131132614 | 0.19 |
ENST00000307968.7 ENST00000307954.8 |
FNIP1 |
folliculin interacting protein 1 |
| chr2_-_27545921 | 0.19 |
ENST00000402310.1 ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17 |
MpV17 mitochondrial inner membrane protein |
| chr15_+_76135622 | 0.19 |
ENST00000338677.4 ENST00000267938.4 ENST00000569423.1 |
UBE2Q2 |
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr6_+_151561506 | 0.18 |
ENST00000253332.1 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr12_-_56123444 | 0.18 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr6_+_31802364 | 0.18 |
ENST00000375640.3 ENST00000375641.2 |
C6orf48 |
chromosome 6 open reading frame 48 |
| chr5_+_43603229 | 0.18 |
ENST00000344920.4 ENST00000512996.2 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr6_-_32811771 | 0.17 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr3_+_45636219 | 0.17 |
ENST00000273317.4 |
LIMD1 |
LIM domains containing 1 |
| chr8_-_124553437 | 0.17 |
ENST00000517956.1 ENST00000443022.2 |
FBXO32 |
F-box protein 32 |
| chr20_-_2821756 | 0.17 |
ENST00000356872.3 ENST00000439542.1 |
PCED1A |
PC-esterase domain containing 1A |
| chr4_-_186347099 | 0.17 |
ENST00000505357.1 ENST00000264689.6 |
UFSP2 |
UFM1-specific peptidase 2 |
| chr8_+_58890917 | 0.16 |
ENST00000522992.1 |
RP11-1112C15.1 |
RP11-1112C15.1 |
| chr6_-_84937314 | 0.16 |
ENST00000257766.4 ENST00000403245.3 |
KIAA1009 |
KIAA1009 |
| chr22_-_43253189 | 0.16 |
ENST00000437119.2 ENST00000429508.2 ENST00000454099.1 ENST00000263245.5 |
ARFGAP3 |
ADP-ribosylation factor GTPase activating protein 3 |
| chr1_+_11866207 | 0.16 |
ENST00000312413.6 ENST00000346436.6 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr6_-_117923520 | 0.16 |
ENST00000368498.2 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
| chr14_-_61190754 | 0.16 |
ENST00000216513.4 |
SIX4 |
SIX homeobox 4 |
| chr21_+_47706537 | 0.15 |
ENST00000397691.1 |
YBEY |
ybeY metallopeptidase (putative) |
| chr7_-_134143841 | 0.15 |
ENST00000285930.4 |
AKR1B1 |
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr6_-_117923610 | 0.15 |
ENST00000535237.1 ENST00000052569.6 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
| chr20_+_31595406 | 0.15 |
ENST00000170150.3 |
BPIFB2 |
BPI fold containing family B, member 2 |
| chr4_+_128651530 | 0.14 |
ENST00000281154.4 |
SLC25A31 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr19_-_9903666 | 0.14 |
ENST00000592587.1 |
ZNF846 |
zinc finger protein 846 |
| chr17_+_40688190 | 0.14 |
ENST00000225927.2 |
NAGLU |
N-acetylglucosaminidase, alpha |
| chr15_-_90645679 | 0.14 |
ENST00000539790.1 ENST00000559482.1 ENST00000330062.3 |
IDH2 |
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr1_-_27240455 | 0.14 |
ENST00000254227.3 |
NR0B2 |
nuclear receptor subfamily 0, group B, member 2 |
| chr7_+_100464760 | 0.14 |
ENST00000200457.4 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr12_-_76953573 | 0.14 |
ENST00000549646.1 ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr2_-_211036051 | 0.14 |
ENST00000418791.1 ENST00000452086.1 ENST00000281772.9 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
| chr13_+_35516390 | 0.13 |
ENST00000540320.1 ENST00000400445.3 ENST00000310336.4 |
NBEA |
neurobeachin |
| chr11_-_6502534 | 0.13 |
ENST00000254584.2 ENST00000525235.1 ENST00000445086.2 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr9_+_37079888 | 0.13 |
ENST00000429493.1 ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1 |
RP11-220I1.1 |
| chr8_-_99129338 | 0.13 |
ENST00000520507.1 |
HRSP12 |
heat-responsive protein 12 |
| chr6_+_127587755 | 0.13 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr5_-_143550241 | 0.13 |
ENST00000522203.1 |
YIPF5 |
Yip1 domain family, member 5 |
| chr1_+_15632231 | 0.12 |
ENST00000375997.4 ENST00000524761.1 ENST00000375995.3 ENST00000401090.2 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr7_+_23749894 | 0.12 |
ENST00000433467.2 |
STK31 |
serine/threonine kinase 31 |
| chr8_+_1772132 | 0.12 |
ENST00000349830.3 ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr16_-_21314360 | 0.12 |
ENST00000219599.3 ENST00000576703.1 |
CRYM |
crystallin, mu |
| chr8_-_134584152 | 0.12 |
ENST00000521180.1 ENST00000517668.1 ENST00000319914.5 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr11_-_6502580 | 0.12 |
ENST00000423813.2 ENST00000396777.3 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr5_+_133984462 | 0.11 |
ENST00000398844.2 ENST00000322887.4 |
SEC24A |
SEC24 family member A |
| chr16_+_333152 | 0.11 |
ENST00000219406.6 ENST00000404312.1 ENST00000456379.1 |
PDIA2 |
protein disulfide isomerase family A, member 2 |
| chr16_-_30134266 | 0.11 |
ENST00000484663.1 ENST00000478356.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr12_-_48152611 | 0.11 |
ENST00000389212.3 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr5_+_125935960 | 0.10 |
ENST00000297540.4 |
PHAX |
phosphorylated adaptor for RNA export |
| chr20_-_57607347 | 0.10 |
ENST00000395663.1 ENST00000395659.1 ENST00000243997.3 |
ATP5E |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr15_+_75660431 | 0.10 |
ENST00000563278.1 |
RP11-817O13.8 |
RP11-817O13.8 |
| chr11_-_102576537 | 0.10 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr7_-_100171270 | 0.10 |
ENST00000538735.1 |
SAP25 |
Sin3A-associated protein, 25kDa |
| chrX_+_48433326 | 0.10 |
ENST00000376755.1 |
RBM3 |
RNA binding motif (RNP1, RRM) protein 3 |
| chr8_-_99129384 | 0.10 |
ENST00000521560.1 ENST00000254878.3 |
HRSP12 |
heat-responsive protein 12 |
| chr22_-_43411106 | 0.10 |
ENST00000453643.1 ENST00000263246.3 ENST00000337959.4 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
| chr12_-_48152853 | 0.10 |
ENST00000171000.4 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr6_-_36953833 | 0.10 |
ENST00000538808.1 ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1 |
mitochondrial carrier 1 |
| chr1_+_249200395 | 0.10 |
ENST00000355360.4 ENST00000329291.5 ENST00000539153.1 |
PGBD2 |
piggyBac transposable element derived 2 |
| chr8_+_22225041 | 0.10 |
ENST00000289952.5 ENST00000524285.1 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
| chr16_-_30134524 | 0.10 |
ENST00000395202.1 ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr19_+_13875316 | 0.09 |
ENST00000319545.8 ENST00000593245.1 ENST00000040663.6 |
MRI1 |
methylthioribose-1-phosphate isomerase 1 |
| chr12_+_6833237 | 0.09 |
ENST00000229251.3 ENST00000539735.1 ENST00000538410.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr14_+_96342729 | 0.09 |
ENST00000504119.1 |
LINC00617 |
long intergenic non-protein coding RNA 617 |
| chr11_-_111957451 | 0.09 |
ENST00000504148.2 ENST00000541231.1 |
TIMM8B |
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr1_+_101361782 | 0.09 |
ENST00000357650.4 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
| chr1_+_145576007 | 0.09 |
ENST00000369298.1 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr1_+_145575980 | 0.09 |
ENST00000393045.2 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr7_-_73133959 | 0.09 |
ENST00000395155.3 ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A |
syntaxin 1A (brain) |
| chr17_-_37309480 | 0.09 |
ENST00000539608.1 |
PLXDC1 |
plexin domain containing 1 |
| chr11_+_65479702 | 0.09 |
ENST00000530446.1 ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5 |
K(lysine) acetyltransferase 5 |
| chr12_-_49318715 | 0.08 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr9_-_125667618 | 0.08 |
ENST00000423239.2 |
RC3H2 |
ring finger and CCCH-type domains 2 |
| chr21_+_45875354 | 0.08 |
ENST00000291592.4 |
LRRC3 |
leucine rich repeat containing 3 |
| chr5_+_112196919 | 0.08 |
ENST00000505459.1 ENST00000282999.3 ENST00000515463.1 |
SRP19 |
signal recognition particle 19kDa |
| chr17_-_2169425 | 0.08 |
ENST00000570606.1 ENST00000354901.4 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr16_-_28074822 | 0.08 |
ENST00000395724.3 ENST00000380898.2 ENST00000447459.2 |
GSG1L |
GSG1-like |
| chr2_-_69614373 | 0.08 |
ENST00000361060.5 ENST00000357308.4 |
GFPT1 |
glutamine--fructose-6-phosphate transaminase 1 |
| chr17_+_49243792 | 0.08 |
ENST00000393183.3 ENST00000393190.1 |
NME1-NME2 |
NME1-NME2 readthrough |
| chr3_+_148709128 | 0.08 |
ENST00000345003.4 ENST00000296048.6 ENST00000483267.1 |
GYG1 |
glycogenin 1 |
| chr12_+_6833437 | 0.07 |
ENST00000534947.1 ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr11_-_100999775 | 0.07 |
ENST00000263463.5 |
PGR |
progesterone receptor |
| chr8_+_22224760 | 0.07 |
ENST00000359741.5 ENST00000520644.1 ENST00000240095.6 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
| chr12_+_121416437 | 0.07 |
ENST00000402929.1 ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A |
HNF1 homeobox A |
| chr1_+_231376941 | 0.07 |
ENST00000436239.1 ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT |
glyceronephosphate O-acyltransferase |
| chr17_+_65821780 | 0.07 |
ENST00000321892.4 ENST00000335221.5 ENST00000306378.6 |
BPTF |
bromodomain PHD finger transcription factor |
| chr9_-_134145880 | 0.07 |
ENST00000372269.3 ENST00000464831.1 |
FAM78A |
family with sequence similarity 78, member A |
| chr1_-_202896310 | 0.07 |
ENST00000367261.3 |
KLHL12 |
kelch-like family member 12 |
| chr20_+_2821340 | 0.07 |
ENST00000380445.3 ENST00000380469.3 |
VPS16 |
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr5_+_122110691 | 0.07 |
ENST00000379516.2 ENST00000505934.1 ENST00000514949.1 |
SNX2 |
sorting nexin 2 |
| chr11_-_69590101 | 0.07 |
ENST00000168712.1 |
FGF4 |
fibroblast growth factor 4 |
| chr5_-_131132658 | 0.07 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr22_+_44568825 | 0.07 |
ENST00000422871.1 |
PARVG |
parvin, gamma |
| chr17_+_74723031 | 0.07 |
ENST00000586200.1 |
METTL23 |
methyltransferase like 23 |
| chr10_-_22292675 | 0.07 |
ENST00000376946.1 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr2_-_15701422 | 0.06 |
ENST00000441750.1 ENST00000281513.5 |
NBAS |
neuroblastoma amplified sequence |
| chr15_+_59063478 | 0.06 |
ENST00000559228.1 ENST00000450403.2 |
FAM63B |
family with sequence similarity 63, member B |
| chr7_+_23749945 | 0.06 |
ENST00000354639.3 ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31 |
serine/threonine kinase 31 |
| chr7_+_148844516 | 0.06 |
ENST00000420008.2 ENST00000475153.1 |
ZNF398 |
zinc finger protein 398 |
| chrX_+_48432892 | 0.06 |
ENST00000376759.3 ENST00000430348.2 |
RBM3 |
RNA binding motif (RNP1, RRM) protein 3 |
| chr17_+_74722912 | 0.06 |
ENST00000589977.1 ENST00000591571.1 ENST00000592849.1 ENST00000586738.1 ENST00000588783.1 ENST00000588563.1 ENST00000586752.1 ENST00000588302.1 ENST00000590964.1 ENST00000341249.6 ENST00000588822.1 |
METTL23 |
methyltransferase like 23 |
| chr15_+_75315896 | 0.06 |
ENST00000342932.3 ENST00000564923.1 ENST00000569562.1 ENST00000568649.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr8_-_144679296 | 0.06 |
ENST00000317198.6 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr1_-_45956822 | 0.06 |
ENST00000372086.3 ENST00000341771.6 |
TESK2 |
testis-specific kinase 2 |
| chr1_-_45956800 | 0.06 |
ENST00000538496.1 |
TESK2 |
testis-specific kinase 2 |
| chr2_+_201936458 | 0.06 |
ENST00000237889.4 |
NDUFB3 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr2_-_132919528 | 0.06 |
ENST00000409867.1 |
ANKRD30BL |
ankyrin repeat domain 30B-like |
| chr17_-_7137582 | 0.06 |
ENST00000575756.1 ENST00000575458.1 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr12_+_131356582 | 0.05 |
ENST00000448750.3 ENST00000541630.1 ENST00000392369.2 ENST00000254675.3 ENST00000535090.1 ENST00000392367.3 |
RAN |
RAN, member RAS oncogene family |
| chr3_+_148709310 | 0.05 |
ENST00000484197.1 ENST00000492285.2 ENST00000461191.1 |
GYG1 |
glycogenin 1 |
| chr7_+_23749767 | 0.05 |
ENST00000355870.3 |
STK31 |
serine/threonine kinase 31 |
| chr5_-_143550159 | 0.05 |
ENST00000448443.2 ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5 |
Yip1 domain family, member 5 |
| chr4_-_18023350 | 0.05 |
ENST00000539056.1 ENST00000382226.5 ENST00000326877.4 |
LCORL |
ligand dependent nuclear receptor corepressor-like |
| chr11_-_777467 | 0.05 |
ENST00000397472.2 ENST00000524550.1 ENST00000319863.8 ENST00000526325.1 ENST00000442059.2 |
PDDC1 |
Parkinson disease 7 domain containing 1 |
| chr11_-_36310958 | 0.05 |
ENST00000532705.1 ENST00000263401.5 ENST00000452374.2 |
COMMD9 |
COMM domain containing 9 |
| chr20_+_61436146 | 0.05 |
ENST00000290291.6 |
OGFR |
opioid growth factor receptor |
| chr9_+_37120536 | 0.05 |
ENST00000336755.5 ENST00000534928.1 ENST00000322831.6 |
ZCCHC7 |
zinc finger, CCHC domain containing 7 |
| chr11_-_61560053 | 0.05 |
ENST00000537328.1 |
TMEM258 |
transmembrane protein 258 |
| chr4_+_40058411 | 0.05 |
ENST00000261435.6 ENST00000515550.1 |
N4BP2 |
NEDD4 binding protein 2 |
| chr17_-_26694979 | 0.05 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr20_-_33413416 | 0.05 |
ENST00000359003.2 |
NCOA6 |
nuclear receptor coactivator 6 |
| chr12_+_48722763 | 0.05 |
ENST00000335017.1 |
H1FNT |
H1 histone family, member N, testis-specific |
| chr19_-_13030071 | 0.05 |
ENST00000293695.7 |
SYCE2 |
synaptonemal complex central element protein 2 |
| chr15_-_98417780 | 0.05 |
ENST00000503874.3 |
LINC00923 |
long intergenic non-protein coding RNA 923 |
| chr20_-_22559211 | 0.05 |
ENST00000564492.1 |
LINC00261 |
long intergenic non-protein coding RNA 261 |
| chrX_-_102941596 | 0.05 |
ENST00000441076.2 ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr12_-_104532062 | 0.05 |
ENST00000240055.3 |
NFYB |
nuclear transcription factor Y, beta |
| chr22_-_35627045 | 0.05 |
ENST00000423311.1 |
CTA-714B7.5 |
CTA-714B7.5 |
| chr19_+_47634039 | 0.05 |
ENST00000597808.1 ENST00000413379.3 ENST00000600706.1 ENST00000540850.1 ENST00000598840.1 ENST00000600753.1 ENST00000270225.7 ENST00000392776.3 |
SAE1 |
SUMO1 activating enzyme subunit 1 |
| chr14_+_100789669 | 0.05 |
ENST00000361529.3 ENST00000557052.1 |
SLC25A47 |
solute carrier family 25, member 47 |
| chr2_+_201676908 | 0.05 |
ENST00000409226.1 ENST00000452790.2 |
BZW1 |
basic leucine zipper and W2 domains 1 |
| chr6_+_158733692 | 0.05 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr17_-_26695013 | 0.05 |
ENST00000555059.2 |
CTB-96E2.2 |
Homeobox protein SEBOX |
| chr1_-_145039771 | 0.05 |
ENST00000493130.2 ENST00000532801.1 ENST00000478649.2 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr19_-_41256207 | 0.05 |
ENST00000598485.2 ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54 |
chromosome 19 open reading frame 54 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 0.6 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.9 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.3 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.1 | 0.3 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.1 | 0.9 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.7 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.7 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.2 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.5 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.2 | GO:0000189 | MAPK import into nucleus(GO:0000189) regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.0 | 0.5 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.4 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 0.3 | GO:0021859 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.0 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.1 | 0.6 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.5 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |