Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for CTCF_CTCFL
Z-value: 0.91
Transcription factors associated with CTCF_CTCFL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
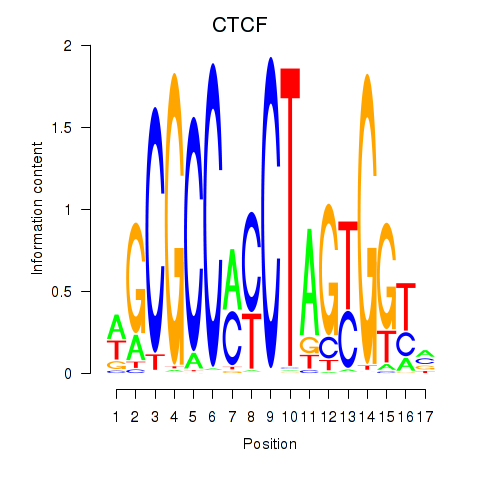
CTCF
|
ENSG00000102974.10 | CTCF |
|
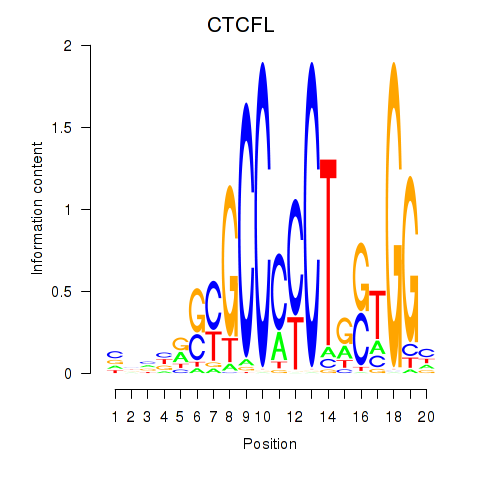
CTCFL
|
ENSG00000124092.8 | CTCFL |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CTCFL | hg19_v2_chr20_-_56100155_56100163 | -0.87 | 5.3e-03 | Click! |
| CTCF | hg19_v2_chr16_+_67596310_67596353 | -0.76 | 2.9e-02 | Click! |
Activity profile of CTCF_CTCFL motif
Sorted Z-values of CTCF_CTCFL motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CTCF_CTCFL
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_111985713 | 0.70 |
ENST00000239007.7 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr14_+_105190514 | 0.66 |
ENST00000330877.2 |
ADSSL1 |
adenylosuccinate synthase like 1 |
| chr11_-_57335280 | 0.65 |
ENST00000287156.4 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
| chr1_-_156675535 | 0.65 |
ENST00000368221.1 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr8_-_48651648 | 0.64 |
ENST00000408965.3 |
CEBPD |
CCAAT/enhancer binding protein (C/EBP), delta |
| chr11_-_72504681 | 0.57 |
ENST00000538536.1 ENST00000543304.1 ENST00000540587.1 ENST00000334805.6 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr19_+_5823813 | 0.56 |
ENST00000303212.2 |
NRTN |
neurturin |
| chr7_+_302918 | 0.56 |
ENST00000599994.1 |
AC187652.1 |
Protein LOC100996433 |
| chr11_+_64781575 | 0.54 |
ENST00000246747.4 ENST00000529384.1 |
ARL2 |
ADP-ribosylation factor-like 2 |
| chr6_+_32821924 | 0.51 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_-_55652290 | 0.49 |
ENST00000589745.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr9_+_130965651 | 0.47 |
ENST00000475805.1 ENST00000341179.7 ENST00000372923.3 |
DNM1 |
dynamin 1 |
| chr9_+_130965677 | 0.46 |
ENST00000393594.3 ENST00000486160.1 |
DNM1 |
dynamin 1 |
| chr7_+_40174565 | 0.45 |
ENST00000309930.5 ENST00000401647.2 ENST00000335693.4 ENST00000413931.1 ENST00000416370.1 ENST00000540834.1 |
C7orf10 |
succinylCoA:glutarate-CoA transferase |
| chr17_-_19266045 | 0.44 |
ENST00000395616.3 |
B9D1 |
B9 protein domain 1 |
| chr1_-_37980344 | 0.44 |
ENST00000448519.2 ENST00000373075.2 ENST00000373073.4 ENST00000296214.5 |
MEAF6 |
MYST/Esa1-associated factor 6 |
| chr14_+_24630465 | 0.43 |
ENST00000557894.1 ENST00000559284.1 ENST00000560275.1 |
IRF9 |
interferon regulatory factor 9 |
| chr19_+_8478154 | 0.42 |
ENST00000381035.4 ENST00000595142.1 ENST00000601724.1 ENST00000393944.1 ENST00000215555.2 ENST00000601283.1 ENST00000595213.1 |
MARCH2 |
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr9_-_140317676 | 0.41 |
ENST00000342129.4 ENST00000340951.4 |
EXD3 |
exonuclease 3'-5' domain containing 3 |
| chr12_-_15374343 | 0.40 |
ENST00000256953.2 ENST00000546331.1 |
RERG |
RAS-like, estrogen-regulated, growth inhibitor |
| chr2_+_130939235 | 0.38 |
ENST00000425361.1 ENST00000457492.1 |
MZT2B |
mitotic spindle organizing protein 2B |
| chr3_+_49209023 | 0.38 |
ENST00000332780.2 |
KLHDC8B |
kelch domain containing 8B |
| chr19_+_46367518 | 0.37 |
ENST00000302177.2 |
FOXA3 |
forkhead box A3 |
| chr9_+_127539481 | 0.37 |
ENST00000373580.3 |
OLFML2A |
olfactomedin-like 2A |
| chr11_+_64781657 | 0.36 |
ENST00000533729.1 |
ARL2 |
ADP-ribosylation factor-like 2 |
| chr9_-_22009241 | 0.36 |
ENST00000380142.4 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr22_-_31503490 | 0.35 |
ENST00000400299.2 |
SELM |
Selenoprotein M |
| chr12_-_7245125 | 0.34 |
ENST00000542285.1 ENST00000540610.1 |
C1R |
complement component 1, r subcomponent |
| chr19_-_49149553 | 0.34 |
ENST00000084798.4 |
CA11 |
carbonic anhydrase XI |
| chr12_+_118814344 | 0.34 |
ENST00000397564.2 |
SUDS3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr12_+_56618102 | 0.34 |
ENST00000267023.4 ENST00000380198.2 ENST00000341463.5 |
NABP2 |
nucleic acid binding protein 2 |
| chr20_+_49411543 | 0.33 |
ENST00000609336.1 ENST00000445038.1 |
BCAS4 |
breast carcinoma amplified sequence 4 |
| chr6_-_30043539 | 0.33 |
ENST00000376751.3 ENST00000244360.6 |
RNF39 |
ring finger protein 39 |
| chr8_-_101571933 | 0.33 |
ENST00000520311.1 |
ANKRD46 |
ankyrin repeat domain 46 |
| chr19_+_49977466 | 0.33 |
ENST00000596435.1 ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr17_-_26220366 | 0.33 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr17_+_43213004 | 0.33 |
ENST00000586346.1 ENST00000398322.3 ENST00000592162.1 ENST00000376955.4 ENST00000321854.8 |
ACBD4 |
acyl-CoA binding domain containing 4 |
| chr5_-_179636153 | 0.32 |
ENST00000361132.4 |
RASGEF1C |
RasGEF domain family, member 1C |
| chr12_+_118814185 | 0.32 |
ENST00000543473.1 |
SUDS3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr20_+_48807351 | 0.32 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr17_-_4890919 | 0.31 |
ENST00000572543.1 ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2 |
calmodulin binding transcription activator 2 |
| chr7_-_6388537 | 0.31 |
ENST00000313324.4 ENST00000530143.1 |
FAM220A |
family with sequence similarity 220, member A |
| chr12_+_50505762 | 0.31 |
ENST00000550487.1 ENST00000317943.2 |
COX14 |
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
| chr10_-_90342947 | 0.31 |
ENST00000437752.1 ENST00000331772.4 |
RNLS |
renalase, FAD-dependent amine oxidase |
| chr19_-_55653259 | 0.30 |
ENST00000593194.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr7_-_100171270 | 0.30 |
ENST00000538735.1 |
SAP25 |
Sin3A-associated protein, 25kDa |
| chr2_+_130939827 | 0.30 |
ENST00000409255.1 ENST00000455239.1 |
MZT2B |
mitotic spindle organizing protein 2B |
| chr17_+_78075361 | 0.30 |
ENST00000577106.1 ENST00000390015.3 |
GAA |
glucosidase, alpha; acid |
| chr12_+_69201923 | 0.30 |
ENST00000462284.1 ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr17_+_78075498 | 0.29 |
ENST00000302262.3 |
GAA |
glucosidase, alpha; acid |
| chr17_-_4890649 | 0.29 |
ENST00000361571.5 |
CAMTA2 |
calmodulin binding transcription activator 2 |
| chr16_+_23569021 | 0.29 |
ENST00000567212.1 ENST00000567264.1 |
UBFD1 |
ubiquitin family domain containing 1 |
| chr12_-_54785074 | 0.29 |
ENST00000338010.5 ENST00000550774.1 |
ZNF385A |
zinc finger protein 385A |
| chr4_+_81256871 | 0.29 |
ENST00000358105.3 ENST00000508675.1 |
C4orf22 |
chromosome 4 open reading frame 22 |
| chr12_+_26274917 | 0.28 |
ENST00000538142.1 |
SSPN |
sarcospan |
| chr9_-_101558777 | 0.28 |
ENST00000375018.1 ENST00000353234.4 |
ANKS6 |
ankyrin repeat and sterile alpha motif domain containing 6 |
| chr19_+_49977818 | 0.28 |
ENST00000594009.1 ENST00000595510.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr12_-_54785054 | 0.27 |
ENST00000352268.6 ENST00000549962.1 |
ZNF385A |
zinc finger protein 385A |
| chr20_+_49411523 | 0.27 |
ENST00000371608.2 |
BCAS4 |
breast carcinoma amplified sequence 4 |
| chr2_-_132249955 | 0.27 |
ENST00000309451.6 |
MZT2A |
mitotic spindle organizing protein 2A |
| chr2_+_233497931 | 0.27 |
ENST00000264059.3 |
EFHD1 |
EF-hand domain family, member D1 |
| chr15_-_66790146 | 0.27 |
ENST00000316634.5 |
SNAPC5 |
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr20_+_32319463 | 0.27 |
ENST00000342427.2 ENST00000375200.1 |
ZNF341 |
zinc finger protein 341 |
| chr12_-_82752159 | 0.26 |
ENST00000552377.1 |
CCDC59 |
coiled-coil domain containing 59 |
| chr2_+_90060377 | 0.26 |
ENST00000436451.2 |
IGKV6D-21 |
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr20_-_30311703 | 0.26 |
ENST00000450273.1 ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1 |
BCL2-like 1 |
| chr2_+_131113609 | 0.26 |
ENST00000347849.3 |
PTPN18 |
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr3_-_52008016 | 0.26 |
ENST00000489595.2 ENST00000461108.1 ENST00000395008.2 ENST00000525795.1 ENST00000361143.5 |
RP11-155D18.14 ABHD14B |
Poly(rC)-binding protein 4 abhydrolase domain containing 14B |
| chr12_+_50505963 | 0.26 |
ENST00000550654.1 ENST00000548985.1 |
COX14 |
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
| chr1_-_109655355 | 0.26 |
ENST00000369945.3 |
C1orf194 |
chromosome 1 open reading frame 194 |
| chr7_+_130126165 | 0.26 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr5_-_76788024 | 0.25 |
ENST00000515253.1 ENST00000414719.2 ENST00000507654.1 ENST00000514559.1 ENST00000511791.1 |
WDR41 |
WD repeat domain 41 |
| chr19_-_38747172 | 0.25 |
ENST00000347262.4 ENST00000591585.1 ENST00000301242.4 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr7_+_130126012 | 0.25 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr5_+_116790848 | 0.24 |
ENST00000504107.1 |
LINC00992 |
long intergenic non-protein coding RNA 992 |
| chr3_-_126373929 | 0.24 |
ENST00000523403.1 ENST00000524230.2 |
TXNRD3 |
thioredoxin reductase 3 |
| chr5_+_75699040 | 0.24 |
ENST00000274364.6 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr2_+_90108504 | 0.24 |
ENST00000390271.2 |
IGKV6D-41 |
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr12_+_7033616 | 0.24 |
ENST00000356654.4 |
ATN1 |
atrophin 1 |
| chr19_+_507299 | 0.24 |
ENST00000359315.5 |
TPGS1 |
tubulin polyglutamylase complex subunit 1 |
| chr9_-_134615443 | 0.24 |
ENST00000372195.1 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr9_-_94712434 | 0.24 |
ENST00000375708.3 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
| chr4_-_140216948 | 0.24 |
ENST00000265500.4 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr9_-_140317605 | 0.24 |
ENST00000479452.1 ENST00000465160.2 |
EXD3 |
exonuclease 3'-5' domain containing 3 |
| chr11_+_67033881 | 0.24 |
ENST00000308595.5 ENST00000526285.1 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
| chr17_-_43209862 | 0.23 |
ENST00000322765.5 |
PLCD3 |
phospholipase C, delta 3 |
| chr1_+_28844778 | 0.23 |
ENST00000411533.1 |
RCC1 |
regulator of chromosome condensation 1 |
| chr5_-_76788317 | 0.23 |
ENST00000296679.4 |
WDR41 |
WD repeat domain 41 |
| chr14_-_103987679 | 0.23 |
ENST00000553610.1 |
CKB |
creatine kinase, brain |
| chr22_+_24204375 | 0.23 |
ENST00000433835.3 |
AP000350.10 |
Uncharacterized protein |
| chr4_+_25314388 | 0.23 |
ENST00000302874.4 |
ZCCHC4 |
zinc finger, CCHC domain containing 4 |
| chr2_-_89459813 | 0.23 |
ENST00000390256.2 |
IGKV6-21 |
immunoglobulin kappa variable 6-21 (non-functional) |
| chr1_+_47799542 | 0.23 |
ENST00000471289.2 ENST00000450808.2 |
CMPK1 |
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr19_+_5690297 | 0.23 |
ENST00000582463.1 ENST00000579446.1 ENST00000394580.2 |
RPL36 |
ribosomal protein L36 |
| chr15_+_63340553 | 0.23 |
ENST00000334895.5 |
TPM1 |
tropomyosin 1 (alpha) |
| chr12_-_102224457 | 0.23 |
ENST00000549165.1 ENST00000549940.1 ENST00000392919.4 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chrX_+_23801280 | 0.23 |
ENST00000379251.3 ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1 |
spermidine/spermine N1-acetyltransferase 1 |
| chr5_-_76788134 | 0.23 |
ENST00000507029.1 |
WDR41 |
WD repeat domain 41 |
| chr19_+_38893751 | 0.23 |
ENST00000588262.1 ENST00000252530.5 ENST00000343358.7 |
FAM98C |
family with sequence similarity 98, member C |
| chr14_-_77608121 | 0.22 |
ENST00000319374.4 |
ZDHHC22 |
zinc finger, DHHC-type containing 22 |
| chr9_-_130742792 | 0.22 |
ENST00000373095.1 |
FAM102A |
family with sequence similarity 102, member A |
| chr4_-_106394866 | 0.22 |
ENST00000502596.1 |
PPA2 |
pyrophosphatase (inorganic) 2 |
| chr6_-_32157947 | 0.22 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr19_-_38746979 | 0.22 |
ENST00000591291.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr10_+_126150369 | 0.22 |
ENST00000392757.4 ENST00000368842.5 ENST00000368839.1 |
LHPP |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr22_+_25003606 | 0.22 |
ENST00000432867.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr2_+_166095898 | 0.21 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr2_-_232395169 | 0.21 |
ENST00000305141.4 |
NMUR1 |
neuromedin U receptor 1 |
| chr22_+_22936998 | 0.21 |
ENST00000390303.2 |
IGLV3-32 |
immunoglobulin lambda variable 3-32 (non-functional) |
| chr20_+_30028322 | 0.21 |
ENST00000376309.3 |
DEFB123 |
defensin, beta 123 |
| chr19_-_2050852 | 0.21 |
ENST00000541165.1 ENST00000591601.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr13_+_88324870 | 0.21 |
ENST00000325089.6 |
SLITRK5 |
SLIT and NTRK-like family, member 5 |
| chr17_-_42441204 | 0.21 |
ENST00000293443.7 |
FAM171A2 |
family with sequence similarity 171, member A2 |
| chr4_-_106395135 | 0.21 |
ENST00000310267.7 |
PPA2 |
pyrophosphatase (inorganic) 2 |
| chr16_+_230435 | 0.21 |
ENST00000199708.2 |
HBQ1 |
hemoglobin, theta 1 |
| chr11_+_8008867 | 0.21 |
ENST00000309828.4 ENST00000449102.2 |
EIF3F |
eukaryotic translation initiation factor 3, subunit F |
| chr19_+_5690207 | 0.21 |
ENST00000347512.3 |
RPL36 |
ribosomal protein L36 |
| chr16_+_1203194 | 0.21 |
ENST00000348261.5 ENST00000358590.4 |
CACNA1H |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr12_-_110486348 | 0.20 |
ENST00000547573.1 ENST00000546651.2 ENST00000551185.2 |
C12orf76 |
chromosome 12 open reading frame 76 |
| chr16_-_54963026 | 0.20 |
ENST00000560208.1 ENST00000557792.1 |
CRNDE |
colorectal neoplasia differentially expressed (non-protein coding) |
| chr2_+_113342011 | 0.20 |
ENST00000324913.5 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr17_+_72199721 | 0.20 |
ENST00000439590.2 ENST00000311111.6 ENST00000584577.1 ENST00000534490.1 ENST00000528433.2 ENST00000533498.1 |
RPL38 |
ribosomal protein L38 |
| chr2_+_113342163 | 0.20 |
ENST00000409719.1 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr19_+_48216600 | 0.20 |
ENST00000263277.3 ENST00000538399.1 |
EHD2 |
EH-domain containing 2 |
| chr17_-_19265982 | 0.20 |
ENST00000268841.6 ENST00000261499.4 ENST00000575478.1 |
B9D1 |
B9 protein domain 1 |
| chr21_-_44495964 | 0.19 |
ENST00000398168.1 ENST00000398165.3 |
CBS |
cystathionine-beta-synthase |
| chr9_-_139922631 | 0.19 |
ENST00000341511.6 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr19_+_42363917 | 0.19 |
ENST00000598742.1 |
RPS19 |
ribosomal protein S19 |
| chr8_+_22436635 | 0.19 |
ENST00000452226.1 ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr3_+_141457030 | 0.19 |
ENST00000273480.3 |
RNF7 |
ring finger protein 7 |
| chr12_-_124018252 | 0.19 |
ENST00000376874.4 |
RILPL1 |
Rab interacting lysosomal protein-like 1 |
| chr11_-_72504637 | 0.19 |
ENST00000536377.1 ENST00000359373.5 |
STARD10 ARAP1 |
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr16_+_67313412 | 0.19 |
ENST00000379344.3 ENST00000568621.1 ENST00000450733.1 ENST00000567938.1 |
PLEKHG4 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr4_+_89378261 | 0.19 |
ENST00000264350.3 |
HERC5 |
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr6_+_3849584 | 0.19 |
ENST00000380274.1 ENST00000380272.3 |
FAM50B |
family with sequence similarity 50, member B |
| chr18_+_596982 | 0.19 |
ENST00000579912.1 ENST00000400606.2 ENST00000540035.1 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr10_-_71906342 | 0.19 |
ENST00000287078.6 ENST00000335494.5 |
TYSND1 |
trypsin domain containing 1 |
| chr10_+_99332529 | 0.19 |
ENST00000455090.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr1_-_40367668 | 0.19 |
ENST00000397332.2 ENST00000429311.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr11_+_117103441 | 0.19 |
ENST00000531287.1 ENST00000531452.1 |
RNF214 |
ring finger protein 214 |
| chr16_+_2867164 | 0.19 |
ENST00000455114.1 ENST00000450020.3 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr22_+_32870661 | 0.19 |
ENST00000266087.7 |
FBXO7 |
F-box protein 7 |
| chr10_+_99332198 | 0.19 |
ENST00000307518.5 ENST00000298808.5 ENST00000370655.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr9_+_140317802 | 0.19 |
ENST00000341349.2 ENST00000392815.2 |
NOXA1 |
NADPH oxidase activator 1 |
| chr3_+_44903361 | 0.19 |
ENST00000302392.4 |
TMEM42 |
transmembrane protein 42 |
| chr6_+_146864829 | 0.18 |
ENST00000367495.3 |
RAB32 |
RAB32, member RAS oncogene family |
| chr1_+_20208870 | 0.18 |
ENST00000375120.3 |
OTUD3 |
OTU domain containing 3 |
| chr19_-_2051223 | 0.18 |
ENST00000309340.7 ENST00000589534.1 ENST00000250896.3 ENST00000589509.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr6_+_125474795 | 0.18 |
ENST00000304877.13 ENST00000534000.1 ENST00000368402.5 ENST00000368388.2 |
TPD52L1 |
tumor protein D52-like 1 |
| chr8_-_101571964 | 0.18 |
ENST00000520552.1 ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46 |
ankyrin repeat domain 46 |
| chr8_-_102217796 | 0.18 |
ENST00000519744.1 ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706 |
zinc finger protein 706 |
| chr9_-_139922726 | 0.18 |
ENST00000265662.5 ENST00000371605.3 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr6_+_125474939 | 0.18 |
ENST00000527711.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr1_+_10490779 | 0.18 |
ENST00000477755.1 |
APITD1 |
apoptosis-inducing, TAF9-like domain 1 |
| chr1_-_146644036 | 0.18 |
ENST00000425272.2 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr11_-_64052111 | 0.18 |
ENST00000394532.3 ENST00000394531.3 ENST00000309032.3 |
BAD |
BCL2-associated agonist of cell death |
| chr17_+_4613776 | 0.18 |
ENST00000269260.2 |
ARRB2 |
arrestin, beta 2 |
| chr19_-_36001286 | 0.18 |
ENST00000602679.1 ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN |
dermokine |
| chr2_+_182756915 | 0.18 |
ENST00000428267.2 |
SSFA2 |
sperm specific antigen 2 |
| chr21_-_44496441 | 0.18 |
ENST00000359624.3 ENST00000352178.5 |
CBS |
cystathionine-beta-synthase |
| chr17_-_45908875 | 0.18 |
ENST00000351111.2 ENST00000414011.1 |
MRPL10 |
mitochondrial ribosomal protein L10 |
| chr1_+_171283331 | 0.17 |
ENST00000367749.3 |
FMO4 |
flavin containing monooxygenase 4 |
| chr14_+_105941118 | 0.17 |
ENST00000550577.1 ENST00000538259.2 |
CRIP2 |
cysteine-rich protein 2 |
| chr8_+_22436248 | 0.17 |
ENST00000308354.7 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr6_+_125475335 | 0.17 |
ENST00000532429.1 ENST00000534199.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr22_+_31608219 | 0.17 |
ENST00000406516.1 ENST00000444929.2 ENST00000331728.4 |
LIMK2 |
LIM domain kinase 2 |
| chr6_+_127588020 | 0.17 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr11_+_61520075 | 0.17 |
ENST00000278836.5 |
MYRF |
myelin regulatory factor |
| chr6_+_125474992 | 0.17 |
ENST00000528193.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr6_-_83775489 | 0.17 |
ENST00000369747.3 |
UBE3D |
ubiquitin protein ligase E3D |
| chr22_-_50765489 | 0.17 |
ENST00000413817.3 |
DENND6B |
DENN/MADD domain containing 6B |
| chr1_+_33207381 | 0.17 |
ENST00000401073.2 |
KIAA1522 |
KIAA1522 |
| chr6_+_127587755 | 0.17 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr17_-_19265855 | 0.17 |
ENST00000440841.1 ENST00000395615.1 ENST00000461069.2 |
B9D1 |
B9 protein domain 1 |
| chr5_+_75699149 | 0.16 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr3_+_141457105 | 0.16 |
ENST00000480908.1 ENST00000393000.3 |
RNF7 |
ring finger protein 7 |
| chr16_-_69788816 | 0.16 |
ENST00000268802.5 |
NOB1 |
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chrX_+_11776410 | 0.16 |
ENST00000361672.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr19_-_48018203 | 0.16 |
ENST00000595227.1 ENST00000593761.1 ENST00000263354.3 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr19_-_4400415 | 0.16 |
ENST00000598564.1 ENST00000417295.2 ENST00000269886.3 |
SH3GL1 |
SH3-domain GRB2-like 1 |
| chr4_-_106395197 | 0.16 |
ENST00000508518.1 ENST00000354147.3 ENST00000432483.2 ENST00000510015.1 ENST00000504028.1 ENST00000348706.5 ENST00000357415.4 ENST00000380004.2 ENST00000341695.5 |
PPA2 |
pyrophosphatase (inorganic) 2 |
| chr7_-_102119342 | 0.16 |
ENST00000393794.3 ENST00000292614.5 |
POLR2J |
polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa |
| chr3_+_101292939 | 0.15 |
ENST00000265260.3 ENST00000469941.1 ENST00000296024.5 |
PCNP |
PEST proteolytic signal containing nuclear protein |
| chr16_+_2867228 | 0.15 |
ENST00000005995.3 ENST00000574813.1 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr11_-_124767693 | 0.15 |
ENST00000533054.1 |
ROBO4 |
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr10_-_103603568 | 0.15 |
ENST00000356640.2 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr19_-_55658281 | 0.15 |
ENST00000585321.2 ENST00000587465.2 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr20_+_49411431 | 0.15 |
ENST00000358791.5 ENST00000262591.5 |
BCAS4 |
breast carcinoma amplified sequence 4 |
| chr18_-_19180681 | 0.15 |
ENST00000269214.5 |
ESCO1 |
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr2_-_64371546 | 0.15 |
ENST00000358912.4 |
PELI1 |
pellino E3 ubiquitin protein ligase 1 |
| chr18_+_616711 | 0.15 |
ENST00000579494.1 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr20_-_36793774 | 0.15 |
ENST00000361475.2 |
TGM2 |
transglutaminase 2 |
| chr1_+_22307592 | 0.15 |
ENST00000400277.2 |
CELA3B |
chymotrypsin-like elastase family, member 3B |
| chr15_-_65715401 | 0.15 |
ENST00000352385.2 |
IGDCC4 |
immunoglobulin superfamily, DCC subclass, member 4 |
| chr3_-_186857267 | 0.15 |
ENST00000455270.1 ENST00000296277.4 |
RPL39L |
ribosomal protein L39-like |
| chr6_+_31105426 | 0.15 |
ENST00000547221.1 |
PSORS1C1 |
psoriasis susceptibility 1 candidate 1 |
| chr19_+_17416457 | 0.14 |
ENST00000252602.1 |
MRPL34 |
mitochondrial ribosomal protein L34 |
| chr18_+_616672 | 0.14 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr16_+_21695750 | 0.14 |
ENST00000388956.4 |
OTOA |
otoancorin |
| chr15_+_63340775 | 0.14 |
ENST00000559281.1 ENST00000317516.7 |
TPM1 |
tropomyosin 1 (alpha) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 0.6 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.6 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 0.6 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 0.7 | GO:1902164 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.4 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 0.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.3 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.3 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.2 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.3 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.2 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.5 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.8 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.4 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.2 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.2 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.2 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.1 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.2 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.0 | 0.2 | GO:0018095 | sperm axoneme assembly(GO:0007288) protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.3 | GO:1902074 | response to salt(GO:1902074) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.7 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.2 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.3 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.1 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.4 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.6 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.0 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) |
| 0.0 | 0.3 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.0 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.3 | GO:1903540 | neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.0 | 0.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.2 | GO:0032308 | regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.0 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.0 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.0 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.0 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.0 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.1 | GO:0099550 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signalling, modulating synaptic transmission(GO:0099550) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.0 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 0.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.0 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 1.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 0.6 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 0.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.4 | GO:0098809 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.1 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.3 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.2 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 1.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.1 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.0 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |