Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
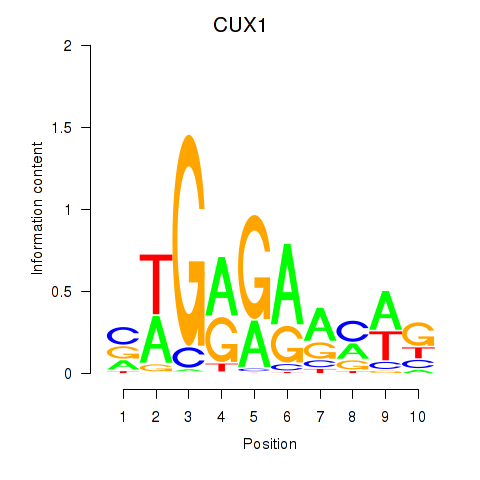
Results for CUX1
Z-value: 1.73
Transcription factors associated with CUX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX1
|
ENSG00000257923.5 | CUX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX1 | hg19_v2_chr7_+_101460882_101460923 | 0.50 | 2.1e-01 | Click! |
Activity profile of CUX1 motif
Sorted Z-values of CUX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153588334 | 3.67 |
ENST00000476873.1 |
S100A14 |
S100 calcium binding protein A14 |
| chr1_-_153588765 | 3.18 |
ENST00000368701.1 ENST00000344616.2 |
S100A14 |
S100 calcium binding protein A14 |
| chr1_+_153003671 | 2.93 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr15_+_40532058 | 2.92 |
ENST00000260404.4 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr12_-_52845910 | 2.89 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr18_-_47376197 | 2.78 |
ENST00000592688.1 |
MYO5B |
myosin VB |
| chr1_-_209824643 | 2.64 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr4_-_15939963 | 2.59 |
ENST00000259988.2 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
| chr15_+_43885252 | 2.48 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr2_-_113594279 | 2.36 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr10_-_116444371 | 2.32 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr20_-_43883197 | 2.13 |
ENST00000338380.2 |
SLPI |
secretory leukocyte peptidase inhibitor |
| chr15_+_40531621 | 2.12 |
ENST00000560346.1 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr8_-_91095099 | 2.12 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chr19_+_35609380 | 2.01 |
ENST00000604621.1 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr15_+_43985725 | 2.01 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr19_-_55658650 | 1.98 |
ENST00000589226.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr16_-_85784634 | 1.98 |
ENST00000284245.4 ENST00000602914.1 |
C16orf74 |
chromosome 16 open reading frame 74 |
| chr19_-_55658281 | 1.93 |
ENST00000585321.2 ENST00000587465.2 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr19_-_55658687 | 1.90 |
ENST00000593046.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr19_-_36004543 | 1.87 |
ENST00000339686.3 ENST00000447113.2 ENST00000440396.1 |
DMKN |
dermokine |
| chr17_+_7942335 | 1.81 |
ENST00000380183.4 ENST00000572022.1 ENST00000380173.2 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr7_+_26191809 | 1.78 |
ENST00000056233.3 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
| chr14_+_21510385 | 1.73 |
ENST00000298690.4 |
RNASE7 |
ribonuclease, RNase A family, 7 |
| chr1_-_207206092 | 1.69 |
ENST00000359470.5 ENST00000461135.2 |
C1orf116 |
chromosome 1 open reading frame 116 |
| chr1_-_209979465 | 1.65 |
ENST00000542854.1 |
IRF6 |
interferon regulatory factor 6 |
| chr1_+_155100342 | 1.46 |
ENST00000368406.2 |
EFNA1 |
ephrin-A1 |
| chr1_+_161676739 | 1.45 |
ENST00000236938.6 ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA |
Fc receptor-like A |
| chr16_-_31147020 | 1.44 |
ENST00000568261.1 ENST00000567797.1 ENST00000317508.6 |
PRSS8 |
protease, serine, 8 |
| chr1_+_161676983 | 1.42 |
ENST00000367957.2 |
FCRLA |
Fc receptor-like A |
| chr1_+_161677034 | 1.42 |
ENST00000349527.4 ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA |
Fc receptor-like A |
| chr5_+_140186647 | 1.37 |
ENST00000512229.2 ENST00000356878.4 ENST00000530339.1 |
PCDHA4 |
protocadherin alpha 4 |
| chr20_+_6748311 | 1.37 |
ENST00000378827.4 |
BMP2 |
bone morphogenetic protein 2 |
| chr19_-_43099070 | 1.37 |
ENST00000244336.5 |
CEACAM8 |
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr9_+_12693336 | 1.36 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr6_+_7541808 | 1.31 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr1_-_183560011 | 1.30 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr17_+_7942424 | 1.25 |
ENST00000573359.1 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr19_-_51523275 | 1.21 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr19_-_51523412 | 1.21 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr7_-_44265492 | 1.17 |
ENST00000425809.1 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr10_+_75670862 | 1.16 |
ENST00000446342.1 ENST00000372764.3 ENST00000372762.4 |
PLAU |
plasminogen activator, urokinase |
| chr18_+_11752040 | 1.12 |
ENST00000423027.3 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr7_+_73245193 | 1.11 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr2_-_31637560 | 1.09 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr14_+_61995722 | 1.06 |
ENST00000556347.1 |
RP11-47I22.4 |
RP11-47I22.4 |
| chr16_+_30751953 | 1.05 |
ENST00000483578.1 |
RP11-2C24.4 |
RP11-2C24.4 |
| chr14_-_23623577 | 1.05 |
ENST00000422941.2 ENST00000453702.1 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr11_+_1860200 | 1.05 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr1_+_15250596 | 1.02 |
ENST00000361144.5 |
KAZN |
kazrin, periplakin interacting protein |
| chr9_+_22446814 | 1.01 |
ENST00000325870.2 |
DMRTA1 |
DMRT-like family A1 |
| chr3_+_111717600 | 1.00 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr19_-_51472222 | 0.98 |
ENST00000376851.3 |
KLK6 |
kallikrein-related peptidase 6 |
| chr7_+_141478242 | 0.97 |
ENST00000247881.2 |
TAS2R4 |
taste receptor, type 2, member 4 |
| chr18_+_47088401 | 0.92 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr19_-_51472031 | 0.92 |
ENST00000391808.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr11_+_45943169 | 0.90 |
ENST00000529052.1 ENST00000531526.1 |
GYLTL1B |
glycosyltransferase-like 1B |
| chrX_-_117107542 | 0.90 |
ENST00000371878.1 |
KLHL13 |
kelch-like family member 13 |
| chr5_-_176057518 | 0.90 |
ENST00000393693.2 |
SNCB |
synuclein, beta |
| chrX_-_117107680 | 0.89 |
ENST00000447671.2 ENST00000262820.3 |
KLHL13 |
kelch-like family member 13 |
| chr11_-_124311054 | 0.80 |
ENST00000328064.2 |
OR8B8 |
olfactory receptor, family 8, subfamily B, member 8 |
| chrX_+_135618258 | 0.78 |
ENST00000440515.1 ENST00000456412.1 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr5_-_176057365 | 0.78 |
ENST00000310112.3 |
SNCB |
synuclein, beta |
| chr12_+_20848282 | 0.77 |
ENST00000545604.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr1_+_206730484 | 0.77 |
ENST00000304534.8 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
| chr12_+_20848377 | 0.77 |
ENST00000540354.1 ENST00000266509.2 ENST00000381552.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr9_-_117568365 | 0.76 |
ENST00000374045.4 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
| chr11_+_60691924 | 0.75 |
ENST00000544065.1 ENST00000453848.2 ENST00000005286.4 |
TMEM132A |
transmembrane protein 132A |
| chr15_-_22448819 | 0.71 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr14_+_75745477 | 0.70 |
ENST00000303562.4 ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr4_-_143227088 | 0.69 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_156119798 | 0.68 |
ENST00000355014.2 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_+_24645865 | 0.68 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_24646002 | 0.68 |
ENST00000356046.2 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr8_+_32406179 | 0.68 |
ENST00000405005.3 |
NRG1 |
neuregulin 1 |
| chr2_-_96811170 | 0.68 |
ENST00000288943.4 |
DUSP2 |
dual specificity phosphatase 2 |
| chr1_+_24645807 | 0.68 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chrX_-_78622805 | 0.67 |
ENST00000373298.2 |
ITM2A |
integral membrane protein 2A |
| chr17_-_34270668 | 0.66 |
ENST00000293274.4 |
LYZL6 |
lysozyme-like 6 |
| chr17_-_34270697 | 0.65 |
ENST00000585556.1 |
LYZL6 |
lysozyme-like 6 |
| chr4_-_143226979 | 0.65 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_-_156786634 | 0.65 |
ENST00000392306.2 ENST00000368199.3 |
SH2D2A |
SH2 domain containing 2A |
| chr15_-_75017711 | 0.62 |
ENST00000567032.1 ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1 |
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr15_-_68498376 | 0.62 |
ENST00000540479.1 ENST00000395465.3 |
CALML4 |
calmodulin-like 4 |
| chr11_+_75479850 | 0.62 |
ENST00000376262.3 ENST00000604733.1 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
| chr10_+_24755416 | 0.61 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chrX_+_48916497 | 0.60 |
ENST00000496529.2 ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120 |
coiled-coil domain containing 120 |
| chr1_+_156117149 | 0.60 |
ENST00000435124.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr19_+_51152702 | 0.60 |
ENST00000425202.1 |
C19orf81 |
chromosome 19 open reading frame 81 |
| chr7_-_142139783 | 0.59 |
ENST00000390374.3 |
TRBV7-6 |
T cell receptor beta variable 7-6 |
| chr19_-_51530916 | 0.57 |
ENST00000594768.1 |
KLK11 |
kallikrein-related peptidase 11 |
| chr19_-_49243845 | 0.56 |
ENST00000222145.4 |
RASIP1 |
Ras interacting protein 1 |
| chr14_+_23842018 | 0.55 |
ENST00000397242.2 ENST00000329715.2 |
IL25 |
interleukin 25 |
| chr2_-_74781061 | 0.55 |
ENST00000264094.3 ENST00000393937.2 ENST00000409986.1 |
LOXL3 |
lysyl oxidase-like 3 |
| chr19_+_8117636 | 0.55 |
ENST00000253451.4 ENST00000315626.4 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr12_+_7060432 | 0.54 |
ENST00000318974.9 ENST00000456013.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr8_-_56685966 | 0.54 |
ENST00000334667.2 |
TMEM68 |
transmembrane protein 68 |
| chr8_-_110620284 | 0.54 |
ENST00000529690.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr12_+_21207503 | 0.54 |
ENST00000545916.1 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr15_-_83953466 | 0.52 |
ENST00000345382.2 |
BNC1 |
basonuclin 1 |
| chr7_+_144052381 | 0.51 |
ENST00000498580.1 ENST00000056217.5 |
ARHGEF5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
| chrX_-_84634737 | 0.51 |
ENST00000262753.4 |
POF1B |
premature ovarian failure, 1B |
| chr20_-_18774614 | 0.51 |
ENST00000412553.1 |
LINC00652 |
long intergenic non-protein coding RNA 652 |
| chr3_-_112218205 | 0.51 |
ENST00000383680.4 |
BTLA |
B and T lymphocyte associated |
| chr7_+_157318477 | 0.51 |
ENST00000444154.1 |
AC006372.1 |
AC006372.1 |
| chr19_-_41220957 | 0.50 |
ENST00000596357.1 ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4 |
aarF domain containing kinase 4 |
| chr1_-_116383322 | 0.50 |
ENST00000429731.1 |
NHLH2 |
nescient helix loop helix 2 |
| chrX_+_102469997 | 0.50 |
ENST00000372695.5 ENST00000372691.3 |
BEX4 |
brain expressed, X-linked 4 |
| chr17_-_43502987 | 0.50 |
ENST00000376922.2 |
ARHGAP27 |
Rho GTPase activating protein 27 |
| chr5_+_55149150 | 0.49 |
ENST00000297015.3 |
IL31RA |
interleukin 31 receptor A |
| chr1_+_156123359 | 0.49 |
ENST00000368284.1 ENST00000368286.2 ENST00000438830.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_+_156123318 | 0.49 |
ENST00000368285.3 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr8_+_75736761 | 0.48 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chrX_+_107288239 | 0.48 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr8_-_56685859 | 0.47 |
ENST00000523423.1 ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68 |
transmembrane protein 68 |
| chr17_-_74533963 | 0.47 |
ENST00000293230.5 |
CYGB |
cytoglobin |
| chr11_-_72385437 | 0.46 |
ENST00000418754.2 ENST00000542969.2 ENST00000334456.5 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr18_-_53069419 | 0.46 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr1_+_46152886 | 0.46 |
ENST00000372025.4 |
TMEM69 |
transmembrane protein 69 |
| chr9_+_71986182 | 0.46 |
ENST00000303068.7 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr14_+_22966642 | 0.45 |
ENST00000390496.1 |
TRAJ41 |
T cell receptor alpha joining 41 |
| chr7_-_142120321 | 0.45 |
ENST00000390377.1 |
TRBV7-7 |
T cell receptor beta variable 7-7 |
| chr6_-_33048483 | 0.45 |
ENST00000419277.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr13_-_46716969 | 0.45 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_+_7055631 | 0.45 |
ENST00000543115.1 ENST00000399448.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr5_-_16936340 | 0.45 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr9_-_96717654 | 0.45 |
ENST00000253968.6 |
BARX1 |
BARX homeobox 1 |
| chr21_-_34915147 | 0.44 |
ENST00000381831.3 ENST00000381839.3 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr3_+_125687987 | 0.43 |
ENST00000514116.1 ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr5_+_180467219 | 0.43 |
ENST00000376841.2 ENST00000327705.9 ENST00000376842.3 |
BTNL9 |
butyrophilin-like 9 |
| chr12_-_55378452 | 0.43 |
ENST00000449076.1 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
| chr17_-_41836153 | 0.42 |
ENST00000301691.2 |
SOST |
sclerostin |
| chr8_+_105235572 | 0.42 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr19_+_42259329 | 0.42 |
ENST00000199764.6 |
CEACAM6 |
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr22_-_37584321 | 0.41 |
ENST00000397110.2 ENST00000337843.2 |
C1QTNF6 |
C1q and tumor necrosis factor related protein 6 |
| chr17_-_77925806 | 0.41 |
ENST00000574241.2 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr1_-_151032040 | 0.41 |
ENST00000540998.1 ENST00000357235.5 |
CDC42SE1 |
CDC42 small effector 1 |
| chr4_+_76481258 | 0.40 |
ENST00000311623.4 ENST00000435974.2 |
C4orf26 |
chromosome 4 open reading frame 26 |
| chr5_+_55149356 | 0.40 |
ENST00000490985.1 ENST00000354961.4 |
IL31RA |
interleukin 31 receptor A |
| chr11_-_58980342 | 0.40 |
ENST00000361050.3 |
MPEG1 |
macrophage expressed 1 |
| chr10_+_103911926 | 0.40 |
ENST00000605788.1 ENST00000405356.1 |
NOLC1 |
nucleolar and coiled-body phosphoprotein 1 |
| chr3_+_57741957 | 0.40 |
ENST00000295951.3 |
SLMAP |
sarcolemma associated protein |
| chr5_+_142149932 | 0.39 |
ENST00000274498.4 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr1_-_235098935 | 0.39 |
ENST00000423175.1 |
RP11-443B7.1 |
RP11-443B7.1 |
| chr17_+_48243352 | 0.39 |
ENST00000344627.6 ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA |
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr8_-_86253888 | 0.38 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr1_-_52870059 | 0.38 |
ENST00000371566.1 |
ORC1 |
origin recognition complex, subunit 1 |
| chr3_-_123512688 | 0.38 |
ENST00000475616.1 |
MYLK |
myosin light chain kinase |
| chr15_-_34659349 | 0.38 |
ENST00000314891.6 |
LPCAT4 |
lysophosphatidylcholine acyltransferase 4 |
| chrX_-_119445306 | 0.37 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chr14_+_75536335 | 0.37 |
ENST00000554763.1 ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chrX_+_107288197 | 0.36 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chrX_-_130037198 | 0.36 |
ENST00000370935.1 ENST00000338144.3 ENST00000394363.1 |
ENOX2 |
ecto-NOX disulfide-thiol exchanger 2 |
| chr14_-_25045446 | 0.36 |
ENST00000216336.2 |
CTSG |
cathepsin G |
| chr1_-_153513170 | 0.36 |
ENST00000368717.2 |
S100A5 |
S100 calcium binding protein A5 |
| chr1_-_94079648 | 0.36 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr11_+_60609537 | 0.36 |
ENST00000227520.5 |
CCDC86 |
coiled-coil domain containing 86 |
| chr1_+_202163015 | 0.36 |
ENST00000367278.3 |
LGR6 |
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr22_-_37545972 | 0.35 |
ENST00000216223.5 |
IL2RB |
interleukin 2 receptor, beta |
| chr22_+_23243156 | 0.35 |
ENST00000390323.2 |
IGLC2 |
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr7_+_93535817 | 0.35 |
ENST00000248572.5 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr19_-_14629224 | 0.35 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr16_-_84538218 | 0.35 |
ENST00000562447.1 ENST00000565765.1 ENST00000535580.1 ENST00000343629.6 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
| chr10_+_50817141 | 0.35 |
ENST00000339797.1 |
CHAT |
choline O-acetyltransferase |
| chrX_-_119445263 | 0.34 |
ENST00000309720.5 |
TMEM255A |
transmembrane protein 255A |
| chr13_-_28674693 | 0.34 |
ENST00000537084.1 ENST00000241453.7 ENST00000380982.4 |
FLT3 |
fms-related tyrosine kinase 3 |
| chr8_-_124054587 | 0.34 |
ENST00000259512.4 |
DERL1 |
derlin 1 |
| chr12_+_7072354 | 0.34 |
ENST00000537269.1 |
U47924.27 |
U47924.27 |
| chr1_+_180897269 | 0.34 |
ENST00000367587.1 |
KIAA1614 |
KIAA1614 |
| chr19_-_10628117 | 0.34 |
ENST00000333430.4 |
S1PR5 |
sphingosine-1-phosphate receptor 5 |
| chr2_+_1417228 | 0.33 |
ENST00000382269.3 ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO |
thyroid peroxidase |
| chr11_-_47870019 | 0.33 |
ENST00000378460.2 |
NUP160 |
nucleoporin 160kDa |
| chr17_-_8027402 | 0.33 |
ENST00000541682.2 ENST00000317814.4 ENST00000577735.1 |
HES7 |
hes family bHLH transcription factor 7 |
| chr6_+_160327974 | 0.33 |
ENST00000252660.4 |
MAS1 |
MAS1 oncogene |
| chr1_+_155006300 | 0.33 |
ENST00000295542.1 ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1 |
DC-STAMP domain containing 1 |
| chr9_+_130478345 | 0.33 |
ENST00000373289.3 ENST00000393748.4 |
TTC16 |
tetratricopeptide repeat domain 16 |
| chr19_+_8117881 | 0.33 |
ENST00000390669.3 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr1_+_212208919 | 0.32 |
ENST00000366991.4 ENST00000542077.1 |
DTL |
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr6_-_53013620 | 0.32 |
ENST00000259803.7 |
GCM1 |
glial cells missing homolog 1 (Drosophila) |
| chr1_-_36948879 | 0.32 |
ENST00000373106.1 ENST00000373104.1 ENST00000373103.1 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
| chr14_+_91580777 | 0.31 |
ENST00000525393.2 ENST00000428926.2 ENST00000517362.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chrX_+_153639856 | 0.31 |
ENST00000426834.1 ENST00000369790.4 ENST00000454722.1 ENST00000350743.4 ENST00000299328.5 ENST00000351413.4 |
TAZ |
tafazzin |
| chr7_+_93535866 | 0.31 |
ENST00000429473.1 ENST00000430875.1 ENST00000428834.1 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr11_+_113779704 | 0.31 |
ENST00000537778.1 |
HTR3B |
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr1_-_116383738 | 0.31 |
ENST00000320238.3 |
NHLH2 |
nescient helix loop helix 2 |
| chr14_-_21490590 | 0.31 |
ENST00000557633.1 |
NDRG2 |
NDRG family member 2 |
| chr12_+_6309963 | 0.31 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr22_-_30925150 | 0.30 |
ENST00000437871.1 |
SEC14L6 |
SEC14-like 6 (S. cerevisiae) |
| chr14_+_32546274 | 0.30 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr6_-_32557610 | 0.30 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr16_+_30935896 | 0.30 |
ENST00000562319.1 ENST00000380310.2 |
FBXL19 |
F-box and leucine-rich repeat protein 19 |
| chr15_+_74908147 | 0.30 |
ENST00000568139.1 ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3 |
CDC-like kinase 3 |
| chr10_+_5238793 | 0.30 |
ENST00000263126.1 |
AKR1C4 |
aldo-keto reductase family 1, member C4 |
| chr9_-_34376851 | 0.30 |
ENST00000297625.7 |
KIAA1161 |
KIAA1161 |
| chr11_-_47869865 | 0.30 |
ENST00000530326.1 ENST00000532747.1 |
NUP160 |
nucleoporin 160kDa |
| chr4_+_15341442 | 0.30 |
ENST00000397700.2 ENST00000295297.4 |
C1QTNF7 |
C1q and tumor necrosis factor related protein 7 |
| chrX_+_70316005 | 0.30 |
ENST00000374259.3 |
FOXO4 |
forkhead box O4 |
| chr11_+_3829691 | 0.30 |
ENST00000278243.4 ENST00000463452.2 ENST00000479072.1 ENST00000496834.2 ENST00000469307.2 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chr15_+_75491213 | 0.30 |
ENST00000360639.2 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr5_+_142149955 | 0.30 |
ENST00000378004.3 |
ARHGAP26 |
Rho GTPase activating protein 26 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.6 | 5.8 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.5 | 1.4 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.4 | 3.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 2.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.4 | 1.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 1.5 | GO:0014028 | notochord formation(GO:0014028) |
| 0.3 | 2.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 1.9 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 1.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.3 | 7.1 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.2 | 1.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 2.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.2 | 0.9 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 0.7 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 0.6 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.2 | 1.6 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.2 | 1.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.6 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.2 | 0.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.9 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.2 | 0.6 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 1.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 5.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.4 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 0.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.4 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.1 | 1.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 1.4 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.4 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.7 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.5 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.6 | GO:0009624 | response to nematode(GO:0009624) |
| 0.1 | 2.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 2.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.7 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.6 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 0.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.4 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 2.3 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.5 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 1.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.3 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 0.2 | GO:0032714 | negative regulation of interleukin-13 production(GO:0032696) negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 1.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.3 | GO:0032489 | aminophospholipid transport(GO:0015917) regulation of Cdc42 protein signal transduction(GO:0032489) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.3 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.3 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.1 | 0.4 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 1.7 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 0.2 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.4 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.3 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.2 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.9 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.1 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.5 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.1 | 1.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.1 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 1.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) |
| 0.0 | 0.3 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 1.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 1.0 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.2 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.2 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 5.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.5 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 1.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.3 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP salvage(GO:0032264) |
| 0.0 | 1.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 1.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 1.7 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 1.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 2.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.4 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.0 | 0.2 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.0 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.3 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.3 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 2.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.3 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.4 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0097466 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0044793 | negative regulation by host of viral process(GO:0044793) negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0071496 | cellular response to external stimulus(GO:0071496) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.2 | GO:0032225 | regulation of synaptic transmission, dopaminergic(GO:0032225) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0042445 | hormone metabolic process(GO:0042445) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.2 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 0.4 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.1 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.0 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 7.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 1.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 3.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.7 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.3 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 2.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 3.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 2.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 1.7 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.5 | 7.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.4 | 2.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 2.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.3 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.2 | 0.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 0.6 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 1.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.6 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 1.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.7 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 0.7 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.6 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 1.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 1.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 1.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.5 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 6.6 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.1 | 0.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 2.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.6 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 1.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.3 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.4 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 1.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.3 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.1 | 1.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 3.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.7 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 2.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 4.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.2 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 4.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.5 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.3 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 1.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 1.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.4 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.5 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 2.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 3.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 5.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 11.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 7.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.5 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 2.6 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 3.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |