Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for DLX3_EVX1_MEOX1
Z-value: 0.60
Transcription factors associated with DLX3_EVX1_MEOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
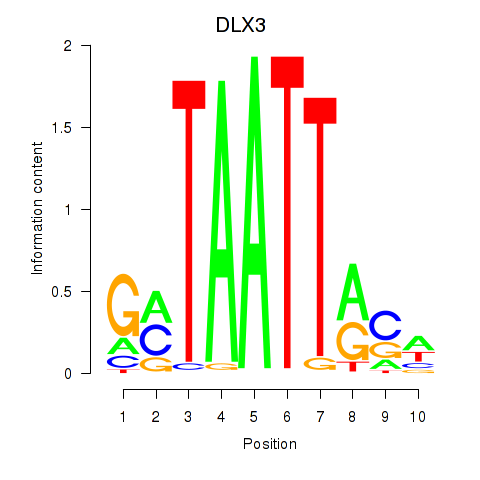
DLX3
|
ENSG00000064195.7 | DLX3 |
|
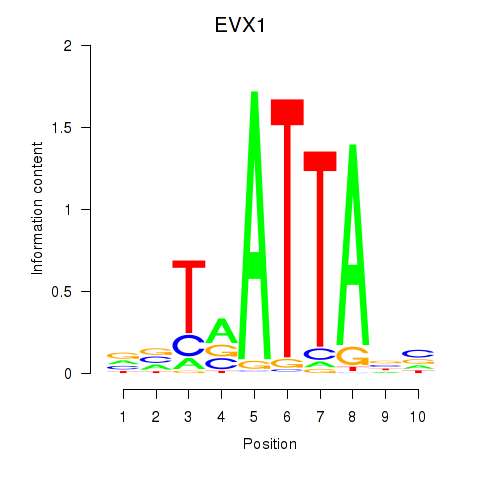
EVX1
|
ENSG00000106038.8 | EVX1 |
|
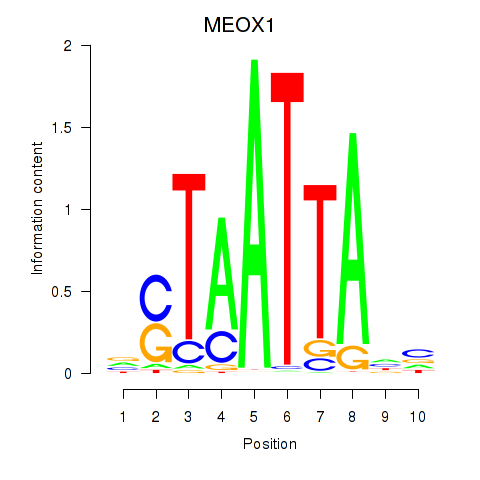
MEOX1
|
ENSG00000005102.8 | MEOX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEOX1 | hg19_v2_chr17_-_41739283_41739322, hg19_v2_chr17_-_41738931_41739040 | -0.32 | 4.4e-01 | Click! |
| DLX3 | hg19_v2_chr17_-_48072574_48072597 | 0.30 | 4.6e-01 | Click! |
| EVX1 | hg19_v2_chr7_+_27282319_27282394 | 0.01 | 9.9e-01 | Click! |
Activity profile of DLX3_EVX1_MEOX1 motif
Sorted Z-values of DLX3_EVX1_MEOX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX3_EVX1_MEOX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_141249154 | 0.35 |
ENST00000357517.5 ENST00000536585.1 |
PCDH1 |
protocadherin 1 |
| chr1_+_160160346 | 0.34 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr1_+_160160283 | 0.33 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr8_-_42234745 | 0.32 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr6_+_29274403 | 0.28 |
ENST00000377160.2 |
OR14J1 |
olfactory receptor, family 14, subfamily J, member 1 |
| chr18_-_33709268 | 0.28 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr4_-_66536057 | 0.26 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr6_+_130339710 | 0.25 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr2_+_169757750 | 0.24 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr5_+_66300446 | 0.23 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_107712173 | 0.22 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr15_+_89631647 | 0.20 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr6_+_151646800 | 0.20 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr15_+_89631381 | 0.19 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr16_+_53133070 | 0.17 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr13_-_46716969 | 0.17 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr22_-_32860427 | 0.16 |
ENST00000534972.1 ENST00000397450.1 ENST00000397452.1 |
BPIFC |
BPI fold containing family C |
| chr10_+_118187379 | 0.16 |
ENST00000369230.3 |
PNLIPRP3 |
pancreatic lipase-related protein 3 |
| chr4_+_77356248 | 0.16 |
ENST00000296043.6 |
SHROOM3 |
shroom family member 3 |
| chr2_+_196313239 | 0.15 |
ENST00000413290.1 |
AC064834.1 |
AC064834.1 |
| chr14_+_22739823 | 0.15 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr7_-_100860851 | 0.15 |
ENST00000223127.3 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr15_+_75080883 | 0.15 |
ENST00000567571.1 |
CSK |
c-src tyrosine kinase |
| chr15_+_41913690 | 0.15 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chr1_-_68698222 | 0.15 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chrX_-_18690210 | 0.14 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
| chr1_-_183622442 | 0.13 |
ENST00000308641.4 |
APOBEC4 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr7_-_33102399 | 0.13 |
ENST00000242210.7 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr16_+_24741013 | 0.13 |
ENST00000315183.7 ENST00000395799.3 |
TNRC6A |
trinucleotide repeat containing 6A |
| chr20_+_60174827 | 0.12 |
ENST00000543233.1 |
CDH4 |
cadherin 4, type 1, R-cadherin (retinal) |
| chr7_-_33102338 | 0.12 |
ENST00000610140.1 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr7_+_116660246 | 0.12 |
ENST00000434836.1 ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr3_-_33686925 | 0.12 |
ENST00000485378.2 ENST00000313350.6 ENST00000487200.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr18_+_32173276 | 0.12 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chr1_+_236958554 | 0.12 |
ENST00000366577.5 ENST00000418145.2 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr14_-_36988882 | 0.11 |
ENST00000498187.2 |
NKX2-1 |
NK2 homeobox 1 |
| chrX_+_130192318 | 0.11 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr7_-_92855762 | 0.11 |
ENST00000453812.2 ENST00000394468.2 |
HEPACAM2 |
HEPACAM family member 2 |
| chr4_-_139163491 | 0.11 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr21_-_32253874 | 0.11 |
ENST00000332378.4 |
KRTAP11-1 |
keratin associated protein 11-1 |
| chr3_+_44840679 | 0.11 |
ENST00000425755.1 |
KIF15 |
kinesin family member 15 |
| chr17_-_18430160 | 0.10 |
ENST00000392176.3 |
FAM106A |
family with sequence similarity 106, member A |
| chr1_-_152386732 | 0.10 |
ENST00000271835.3 |
CRNN |
cornulin |
| chr1_-_197115818 | 0.10 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr17_-_39306054 | 0.10 |
ENST00000343246.4 |
KRTAP4-5 |
keratin associated protein 4-5 |
| chr5_+_98109322 | 0.10 |
ENST00000513185.1 |
RGMB |
repulsive guidance molecule family member b |
| chr7_-_81399411 | 0.10 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chrX_-_19689106 | 0.10 |
ENST00000379716.1 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr8_-_139926236 | 0.10 |
ENST00000303045.6 ENST00000435777.1 |
COL22A1 |
collagen, type XXII, alpha 1 |
| chr3_+_111717600 | 0.09 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr8_+_32579341 | 0.09 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr3_-_123339343 | 0.09 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chrM_+_12331 | 0.09 |
ENST00000361567.2 |
MT-ND5 |
mitochondrially encoded NADH dehydrogenase 5 |
| chr4_-_74486217 | 0.09 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_174451370 | 0.09 |
ENST00000359562.4 |
HAND2 |
heart and neural crest derivatives expressed 2 |
| chr16_-_52640834 | 0.09 |
ENST00000510238.3 |
CASC16 |
cancer susceptibility candidate 16 (non-protein coding) |
| chr1_+_107683436 | 0.09 |
ENST00000370068.1 |
NTNG1 |
netrin G1 |
| chr17_-_46716647 | 0.09 |
ENST00000608940.1 |
RP11-357H14.17 |
RP11-357H14.17 |
| chr2_-_227050079 | 0.09 |
ENST00000423838.1 |
AC068138.1 |
AC068138.1 |
| chr2_+_27498289 | 0.09 |
ENST00000296097.3 ENST00000420191.1 |
DNAJC5G |
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr2_-_31361543 | 0.09 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr3_-_33686743 | 0.09 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr13_-_36050819 | 0.09 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr10_-_105845674 | 0.09 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr19_-_7968427 | 0.08 |
ENST00000539278.1 |
AC010336.1 |
Uncharacterized protein |
| chr6_+_47666275 | 0.08 |
ENST00000327753.3 ENST00000283303.2 |
GPR115 |
G protein-coupled receptor 115 |
| chr12_-_10978957 | 0.08 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr7_-_83824169 | 0.08 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_+_111718036 | 0.08 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr11_-_121986923 | 0.08 |
ENST00000560104.1 |
BLID |
BH3-like motif containing, cell death inducer |
| chr2_-_68052694 | 0.08 |
ENST00000457448.1 |
AC010987.6 |
AC010987.6 |
| chr17_-_39677971 | 0.07 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr3_+_111717511 | 0.07 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr17_-_38911580 | 0.07 |
ENST00000312150.4 |
KRT25 |
keratin 25 |
| chr1_+_107683644 | 0.07 |
ENST00000370067.1 |
NTNG1 |
netrin G1 |
| chr12_-_56694142 | 0.07 |
ENST00000550655.1 ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS |
citrate synthase |
| chr2_-_216003127 | 0.07 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr14_+_22748980 | 0.07 |
ENST00000390465.2 |
TRAV38-2DV8 |
T cell receptor alpha variable 38-2/delta variable 8 |
| chr7_+_73245193 | 0.07 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr1_+_202317855 | 0.07 |
ENST00000356764.2 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr2_+_113299990 | 0.07 |
ENST00000537335.1 ENST00000417433.2 |
POLR1B |
polymerase (RNA) I polypeptide B, 128kDa |
| chr5_-_16509101 | 0.07 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr4_+_88754113 | 0.07 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chrM_+_8527 | 0.07 |
ENST00000361899.2 |
MT-ATP6 |
mitochondrially encoded ATP synthase 6 |
| chrX_-_110655306 | 0.07 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr5_+_126984710 | 0.07 |
ENST00000379445.3 |
CTXN3 |
cortexin 3 |
| chr12_-_52946923 | 0.07 |
ENST00000267119.5 |
KRT71 |
keratin 71 |
| chr11_-_107729887 | 0.07 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr1_-_68698197 | 0.06 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr7_-_73038822 | 0.06 |
ENST00000414749.2 ENST00000429400.2 ENST00000434326.1 |
MLXIPL |
MLX interacting protein-like |
| chr14_-_78083112 | 0.06 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr7_-_73038867 | 0.06 |
ENST00000313375.3 ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL |
MLX interacting protein-like |
| chr4_+_88754069 | 0.06 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr20_-_50418972 | 0.06 |
ENST00000395997.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr11_+_65554493 | 0.06 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr2_-_207082748 | 0.06 |
ENST00000407325.2 ENST00000411719.1 |
GPR1 |
G protein-coupled receptor 1 |
| chr4_-_74486109 | 0.06 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_+_31895254 | 0.06 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chr5_-_87516448 | 0.06 |
ENST00000511218.1 |
TMEM161B |
transmembrane protein 161B |
| chrX_+_105855160 | 0.06 |
ENST00000372544.2 ENST00000372548.4 |
CXorf57 |
chromosome X open reading frame 57 |
| chr4_+_26862400 | 0.06 |
ENST00000467011.1 ENST00000412829.2 |
STIM2 |
stromal interaction molecule 2 |
| chr14_-_82000140 | 0.06 |
ENST00000555824.1 ENST00000557372.1 ENST00000336735.4 |
SEL1L |
sel-1 suppressor of lin-12-like (C. elegans) |
| chr17_+_42264556 | 0.06 |
ENST00000319511.6 ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr1_-_190446759 | 0.06 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr2_-_17981462 | 0.06 |
ENST00000402989.1 ENST00000428868.1 |
SMC6 |
structural maintenance of chromosomes 6 |
| chr12_+_41831485 | 0.06 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr7_-_32338917 | 0.06 |
ENST00000396193.1 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr14_+_64680854 | 0.06 |
ENST00000458046.2 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
| chr8_-_87755878 | 0.06 |
ENST00000320005.5 |
CNGB3 |
cyclic nucleotide gated channel beta 3 |
| chr4_-_116034979 | 0.06 |
ENST00000264363.2 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr4_+_40198527 | 0.06 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr13_-_110438914 | 0.06 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chr4_-_74486347 | 0.06 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr22_-_41682172 | 0.06 |
ENST00000356244.3 |
RANGAP1 |
Ran GTPase activating protein 1 |
| chr3_+_111718173 | 0.06 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr14_+_61654271 | 0.05 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr3_-_74570291 | 0.05 |
ENST00000263665.6 |
CNTN3 |
contactin 3 (plasmacytoma associated) |
| chr2_-_182545603 | 0.05 |
ENST00000295108.3 |
NEUROD1 |
neuronal differentiation 1 |
| chr20_-_50418947 | 0.05 |
ENST00000371539.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr3_-_108248169 | 0.05 |
ENST00000273353.3 |
MYH15 |
myosin, heavy chain 15 |
| chr7_+_129906660 | 0.05 |
ENST00000222481.4 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
| chr12_+_81110684 | 0.05 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr13_+_36050881 | 0.05 |
ENST00000537702.1 |
NBEA |
neurobeachin |
| chr10_-_73848086 | 0.05 |
ENST00000536168.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr1_-_234667504 | 0.05 |
ENST00000421207.1 ENST00000435574.1 |
RP5-855F14.1 |
RP5-855F14.1 |
| chr19_+_11071546 | 0.05 |
ENST00000358026.2 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chrX_+_78426469 | 0.05 |
ENST00000276077.1 |
GPR174 |
G protein-coupled receptor 174 |
| chr17_-_27418537 | 0.05 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr3_+_130569429 | 0.05 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr15_+_63188009 | 0.05 |
ENST00000557900.1 |
RP11-1069G10.2 |
RP11-1069G10.2 |
| chr8_+_50824233 | 0.05 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr17_-_64225508 | 0.05 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr18_+_55888767 | 0.05 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_+_20620946 | 0.04 |
ENST00000525748.1 |
SLC6A5 |
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr15_-_68497657 | 0.04 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr6_-_76203454 | 0.04 |
ENST00000237172.7 |
FILIP1 |
filamin A interacting protein 1 |
| chrX_+_135730297 | 0.04 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chr8_-_103425047 | 0.04 |
ENST00000520539.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr1_+_62439037 | 0.04 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr4_+_174089904 | 0.04 |
ENST00000265000.4 |
GALNT7 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr21_-_43346790 | 0.04 |
ENST00000329623.7 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
| chr19_+_13049413 | 0.04 |
ENST00000316448.5 ENST00000588454.1 |
CALR |
calreticulin |
| chr15_-_64673665 | 0.04 |
ENST00000300035.4 |
KIAA0101 |
KIAA0101 |
| chr6_+_34204642 | 0.04 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr11_-_33913708 | 0.04 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr8_-_110346614 | 0.04 |
ENST00000239690.4 |
NUDCD1 |
NudC domain containing 1 |
| chr8_-_103424916 | 0.04 |
ENST00000220959.4 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr6_-_76203345 | 0.04 |
ENST00000393004.2 |
FILIP1 |
filamin A interacting protein 1 |
| chr12_-_89746173 | 0.04 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr4_+_70796784 | 0.04 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr2_-_99279928 | 0.04 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr4_-_39979576 | 0.04 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr6_-_116833500 | 0.04 |
ENST00000356128.4 |
TRAPPC3L |
trafficking protein particle complex 3-like |
| chr20_-_50419055 | 0.04 |
ENST00000217086.4 |
SALL4 |
spalt-like transcription factor 4 |
| chr16_-_66864806 | 0.04 |
ENST00000566336.1 ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
| chrM_+_7586 | 0.04 |
ENST00000361739.1 |
MT-CO2 |
mitochondrially encoded cytochrome c oxidase II |
| chr19_-_7698599 | 0.04 |
ENST00000311069.5 |
PCP2 |
Purkinje cell protein 2 |
| chr13_+_73632897 | 0.04 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chrX_+_133930798 | 0.04 |
ENST00000414371.2 |
FAM122C |
family with sequence similarity 122C |
| chr9_-_136933615 | 0.04 |
ENST00000371834.2 |
BRD3 |
bromodomain containing 3 |
| chr13_-_31191642 | 0.04 |
ENST00000405805.1 |
HMGB1 |
high mobility group box 1 |
| chr8_-_103424986 | 0.04 |
ENST00000521922.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr20_+_18488137 | 0.04 |
ENST00000450074.1 ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B |
Sec23 homolog B (S. cerevisiae) |
| chr6_-_109702885 | 0.03 |
ENST00000504373.1 |
CD164 |
CD164 molecule, sialomucin |
| chr8_-_82395461 | 0.03 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chr14_+_22984601 | 0.03 |
ENST00000390509.1 |
TRAJ28 |
T cell receptor alpha joining 28 |
| chr10_-_33623310 | 0.03 |
ENST00000395995.1 ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1 |
neuropilin 1 |
| chr2_-_55647057 | 0.03 |
ENST00000436346.1 |
CCDC88A |
coiled-coil domain containing 88A |
| chr9_-_3469181 | 0.03 |
ENST00000366116.2 |
AL365202.1 |
Uncharacterized protein |
| chr1_+_230193521 | 0.03 |
ENST00000543760.1 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr2_-_230786619 | 0.03 |
ENST00000389045.3 ENST00000409677.1 |
TRIP12 |
thyroid hormone receptor interactor 12 |
| chr16_-_67517716 | 0.03 |
ENST00000290953.2 |
AGRP |
agouti related protein homolog (mouse) |
| chr3_-_123680246 | 0.03 |
ENST00000488653.2 |
CCDC14 |
coiled-coil domain containing 14 |
| chr11_-_129062093 | 0.03 |
ENST00000310343.9 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr1_-_201140673 | 0.03 |
ENST00000367333.2 |
TMEM9 |
transmembrane protein 9 |
| chr9_+_125132803 | 0.03 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_-_243326612 | 0.03 |
ENST00000492145.1 ENST00000490813.1 ENST00000464936.1 |
CEP170 |
centrosomal protein 170kDa |
| chr8_+_7783859 | 0.03 |
ENST00000400120.3 |
ZNF705B |
zinc finger protein 705B |
| chr4_-_119759795 | 0.03 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr16_-_2837949 | 0.03 |
ENST00000570702.1 |
PRSS33 |
protease, serine, 33 |
| chr2_-_230786679 | 0.03 |
ENST00000543084.1 ENST00000343290.5 ENST00000389044.4 ENST00000283943.5 |
TRIP12 |
thyroid hormone receptor interactor 12 |
| chr5_-_141030943 | 0.03 |
ENST00000522783.1 ENST00000519800.1 ENST00000435817.2 |
FCHSD1 |
FCH and double SH3 domains 1 |
| chr19_-_47137942 | 0.03 |
ENST00000300873.4 |
GNG8 |
guanine nucleotide binding protein (G protein), gamma 8 |
| chr14_-_21737610 | 0.03 |
ENST00000320084.7 ENST00000449098.1 ENST00000336053.6 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr4_-_25865159 | 0.03 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chrX_-_133931164 | 0.03 |
ENST00000370790.1 ENST00000298090.6 |
FAM122B |
family with sequence similarity 122B |
| chr3_-_195538760 | 0.03 |
ENST00000475231.1 |
MUC4 |
mucin 4, cell surface associated |
| chr5_-_42811986 | 0.03 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr2_-_55646957 | 0.03 |
ENST00000263630.8 |
CCDC88A |
coiled-coil domain containing 88A |
| chr12_-_14849470 | 0.03 |
ENST00000261170.3 |
GUCY2C |
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr20_-_271304 | 0.03 |
ENST00000400269.3 ENST00000360321.2 |
C20orf96 |
chromosome 20 open reading frame 96 |
| chrX_-_106362013 | 0.03 |
ENST00000372487.1 ENST00000372479.3 ENST00000203616.8 |
RBM41 |
RNA binding motif protein 41 |
| chr17_-_54893250 | 0.03 |
ENST00000397862.2 |
C17orf67 |
chromosome 17 open reading frame 67 |
| chr9_+_116225999 | 0.03 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr10_-_101690650 | 0.03 |
ENST00000543621.1 |
DNMBP |
dynamin binding protein |
| chr20_+_30697298 | 0.03 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr1_+_68150744 | 0.03 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chr20_+_43990576 | 0.03 |
ENST00000372727.1 ENST00000414310.1 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr15_+_96869165 | 0.03 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_113763031 | 0.03 |
ENST00000259211.6 |
IL36A |
interleukin 36, alpha |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0061032 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |