Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for DLX5
Z-value: 0.52
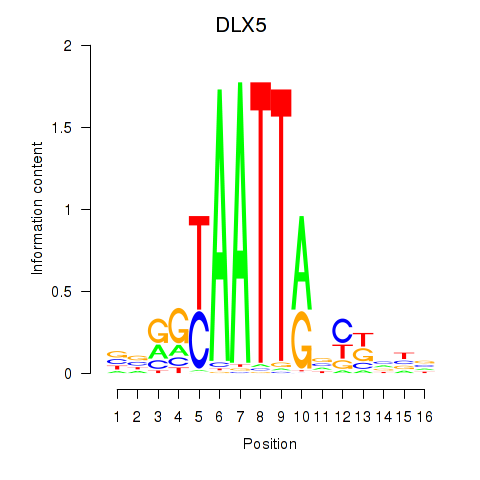
Transcription factors associated with DLX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX5
|
ENSG00000105880.4 | DLX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX5 | hg19_v2_chr7_-_96654133_96654409 | -0.68 | 6.4e-02 | Click! |
Activity profile of DLX5 motif
Sorted Z-values of DLX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_53133070 | 0.86 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr15_+_89631381 | 0.71 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr3_+_186560462 | 0.51 |
ENST00000412955.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr3_+_186560476 | 0.51 |
ENST00000320741.2 ENST00000444204.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr15_+_89631647 | 0.51 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr5_-_20575959 | 0.44 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr12_-_10151773 | 0.32 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr3_-_105588231 | 0.32 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr17_+_42264556 | 0.31 |
ENST00000319511.6 ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr7_+_100770328 | 0.31 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr15_+_41913690 | 0.30 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chr7_+_99717230 | 0.23 |
ENST00000262932.3 |
CNPY4 |
canopy FGF signaling regulator 4 |
| chr7_+_70597109 | 0.23 |
ENST00000333538.5 |
WBSCR17 |
Williams-Beuren syndrome chromosome region 17 |
| chr3_-_105587879 | 0.21 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr12_+_51818749 | 0.21 |
ENST00000514353.3 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr1_+_160160283 | 0.20 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr1_+_160160346 | 0.19 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr3_+_11178779 | 0.18 |
ENST00000438284.2 |
HRH1 |
histamine receptor H1 |
| chr17_+_59489112 | 0.17 |
ENST00000335108.2 |
C17orf82 |
chromosome 17 open reading frame 82 |
| chr3_+_158519654 | 0.17 |
ENST00000415822.2 ENST00000392813.4 ENST00000264266.8 |
MFSD1 |
major facilitator superfamily domain containing 1 |
| chr4_-_74853897 | 0.17 |
ENST00000296028.3 |
PPBP |
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr6_-_78173490 | 0.17 |
ENST00000369947.2 |
HTR1B |
5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
| chr8_-_124553437 | 0.15 |
ENST00000517956.1 ENST00000443022.2 |
FBXO32 |
F-box protein 32 |
| chr2_-_183387430 | 0.15 |
ENST00000410103.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr5_-_160279207 | 0.13 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr13_+_36050881 | 0.13 |
ENST00000537702.1 |
NBEA |
neurobeachin |
| chr10_+_99205959 | 0.13 |
ENST00000352634.4 ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr1_+_202317855 | 0.12 |
ENST00000356764.2 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr5_-_42811986 | 0.12 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr10_+_99205894 | 0.12 |
ENST00000370854.3 ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr11_-_33913708 | 0.11 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chrX_-_16887963 | 0.11 |
ENST00000380084.4 |
RBBP7 |
retinoblastoma binding protein 7 |
| chr6_+_123038689 | 0.11 |
ENST00000354275.2 ENST00000368446.1 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr4_-_138453559 | 0.11 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr2_+_95963052 | 0.11 |
ENST00000295225.5 |
KCNIP3 |
Kv channel interacting protein 3, calsenilin |
| chr1_-_114447412 | 0.11 |
ENST00000369567.1 ENST00000369566.3 |
AP4B1 |
adaptor-related protein complex 4, beta 1 subunit |
| chr17_+_28268623 | 0.11 |
ENST00000394835.3 ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5 |
EF-hand calcium binding domain 5 |
| chr7_-_14028488 | 0.11 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr7_-_14029283 | 0.10 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr7_-_7575477 | 0.10 |
ENST00000399429.3 |
COL28A1 |
collagen, type XXVIII, alpha 1 |
| chr1_-_150669604 | 0.10 |
ENST00000427665.1 ENST00000540514.1 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr19_-_14785674 | 0.10 |
ENST00000253673.5 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr19_-_14785622 | 0.09 |
ENST00000443157.2 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr14_+_22739823 | 0.09 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr2_+_105050794 | 0.09 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr18_-_33709268 | 0.09 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr14_-_68283291 | 0.08 |
ENST00000555452.1 ENST00000347230.4 |
ZFYVE26 |
zinc finger, FYVE domain containing 26 |
| chr2_+_196313239 | 0.08 |
ENST00000413290.1 |
AC064834.1 |
AC064834.1 |
| chr11_-_26593779 | 0.08 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr19_-_14785698 | 0.08 |
ENST00000344373.4 ENST00000595472.1 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr17_-_47785504 | 0.08 |
ENST00000514907.1 ENST00000503334.1 ENST00000508520.1 |
SLC35B1 |
solute carrier family 35, member B1 |
| chr19_+_58570605 | 0.08 |
ENST00000359978.6 ENST00000401053.4 ENST00000439855.2 ENST00000313434.5 ENST00000511556.1 ENST00000506786.1 |
ZNF135 |
zinc finger protein 135 |
| chr11_-_26593677 | 0.08 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr22_-_32860427 | 0.07 |
ENST00000534972.1 ENST00000397450.1 ENST00000397452.1 |
BPIFC |
BPI fold containing family C |
| chr3_+_186288454 | 0.07 |
ENST00000265028.3 |
DNAJB11 |
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr6_-_27860956 | 0.07 |
ENST00000359611.2 |
HIST1H2AM |
histone cluster 1, H2am |
| chr4_-_66536057 | 0.07 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr2_+_228736321 | 0.07 |
ENST00000309931.2 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
| chr13_-_36050819 | 0.07 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr2_+_228736335 | 0.06 |
ENST00000440997.1 ENST00000545118.1 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
| chr11_+_76092353 | 0.06 |
ENST00000530460.1 ENST00000321844.4 |
RP11-111M22.2 |
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr2_+_169757750 | 0.06 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr1_-_197115818 | 0.05 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr2_+_228735763 | 0.05 |
ENST00000373666.2 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
| chr3_+_2933893 | 0.05 |
ENST00000397459.2 |
CNTN4 |
contactin 4 |
| chr11_-_26743546 | 0.05 |
ENST00000280467.6 ENST00000396005.3 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr12_-_10978957 | 0.05 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr3_-_186288097 | 0.05 |
ENST00000446782.1 |
TBCCD1 |
TBCC domain containing 1 |
| chr1_-_23670813 | 0.05 |
ENST00000374612.1 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
| chr15_+_63188009 | 0.05 |
ENST00000557900.1 |
RP11-1069G10.2 |
RP11-1069G10.2 |
| chr14_+_24540046 | 0.05 |
ENST00000397016.2 ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6 |
copine VI (neuronal) |
| chr11_-_102651343 | 0.04 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr19_-_19302931 | 0.04 |
ENST00000444486.3 ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B MEF2BNB MEF2B |
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr15_+_89164520 | 0.04 |
ENST00000332810.3 |
AEN |
apoptosis enhancing nuclease |
| chr8_-_7332604 | 0.04 |
ENST00000316169.2 |
DEFB104B |
defensin, beta 104B |
| chr2_-_183387064 | 0.04 |
ENST00000536095.1 ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_26593649 | 0.04 |
ENST00000455601.2 |
MUC15 |
mucin 15, cell surface associated |
| chr11_+_818902 | 0.04 |
ENST00000336615.4 |
PNPLA2 |
patatin-like phospholipase domain containing 2 |
| chr1_-_23670752 | 0.04 |
ENST00000302271.6 ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
| chr1_+_202317815 | 0.04 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr21_-_32253874 | 0.03 |
ENST00000332378.4 |
KRTAP11-1 |
keratin associated protein 11-1 |
| chr2_-_183387283 | 0.03 |
ENST00000435564.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr21_-_15918618 | 0.03 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr6_-_33160231 | 0.03 |
ENST00000395194.1 ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2 |
collagen, type XI, alpha 2 |
| chr13_-_36788718 | 0.03 |
ENST00000317764.6 ENST00000379881.3 |
SOHLH2 |
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr17_-_39324424 | 0.03 |
ENST00000391356.2 |
KRTAP4-3 |
keratin associated protein 4-3 |
| chr8_+_32579341 | 0.02 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr19_+_36606354 | 0.02 |
ENST00000589996.1 ENST00000591296.1 |
TBCB |
tubulin folding cofactor B |
| chr7_-_99716952 | 0.02 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr5_+_63802109 | 0.02 |
ENST00000334025.2 |
RGS7BP |
regulator of G-protein signaling 7 binding protein |
| chr4_-_105416039 | 0.01 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr1_-_201140673 | 0.01 |
ENST00000367333.2 |
TMEM9 |
transmembrane protein 9 |
| chr22_-_30960876 | 0.01 |
ENST00000401975.1 ENST00000428682.1 ENST00000423299.1 |
GAL3ST1 |
galactose-3-O-sulfotransferase 1 |
| chr7_-_86849883 | 0.01 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr12_+_52695617 | 0.01 |
ENST00000293525.5 |
KRT86 |
keratin 86 |
| chr4_-_8873531 | 0.01 |
ENST00000400677.3 |
HMX1 |
H6 family homeobox 1 |
| chr1_-_150669500 | 0.01 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr19_+_36605850 | 0.00 |
ENST00000221855.3 |
TBCB |
tubulin folding cofactor B |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.3 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.5 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 1.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |