Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for EGR3_EGR2
Z-value: 0.77
Transcription factors associated with EGR3_EGR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
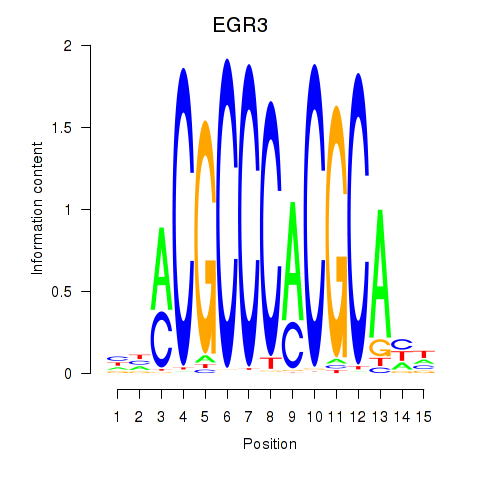
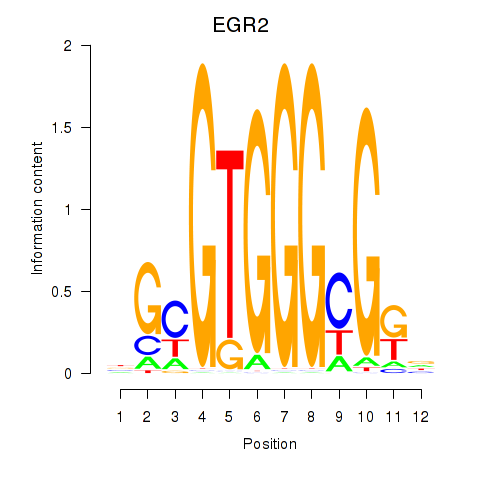
EGR3
|
ENSG00000179388.8 | EGR3 |
|
EGR2
|
ENSG00000122877.9 | EGR2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR3 | hg19_v2_chr8_-_22550815_22550844 | 0.65 | 7.8e-02 | Click! |
| EGR2 | hg19_v2_chr10_-_64576105_64576133 | 0.43 | 2.8e-01 | Click! |
Activity profile of EGR3_EGR2 motif
Sorted Z-values of EGR3_EGR2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR3_EGR2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_100199800 | 0.62 |
ENST00000223061.5 |
PCOLCE |
procollagen C-endopeptidase enhancer |
| chrX_+_16964794 | 0.58 |
ENST00000357277.3 |
REPS2 |
RALBP1 associated Eps domain containing 2 |
| chr6_+_72596406 | 0.55 |
ENST00000491071.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chrX_+_16964985 | 0.51 |
ENST00000303843.7 |
REPS2 |
RALBP1 associated Eps domain containing 2 |
| chr6_+_72596604 | 0.49 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr18_-_500692 | 0.48 |
ENST00000400256.3 |
COLEC12 |
collectin sub-family member 12 |
| chr1_+_215256467 | 0.41 |
ENST00000391894.2 ENST00000444842.2 |
KCNK2 |
potassium channel, subfamily K, member 2 |
| chrX_-_142722897 | 0.40 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr20_-_3996165 | 0.39 |
ENST00000545616.2 ENST00000358395.6 |
RNF24 |
ring finger protein 24 |
| chr7_-_100493482 | 0.39 |
ENST00000411582.1 ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr4_+_145567297 | 0.38 |
ENST00000434550.2 |
HHIP |
hedgehog interacting protein |
| chr13_+_88324870 | 0.37 |
ENST00000325089.6 |
SLITRK5 |
SLIT and NTRK-like family, member 5 |
| chr11_-_2170786 | 0.35 |
ENST00000300632.5 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr17_-_66597530 | 0.35 |
ENST00000592554.1 |
FAM20A |
family with sequence similarity 20, member A |
| chr4_+_145567173 | 0.34 |
ENST00000296575.3 |
HHIP |
hedgehog interacting protein |
| chr4_+_150999418 | 0.34 |
ENST00000296550.7 |
DCLK2 |
doublecortin-like kinase 2 |
| chr2_-_98612350 | 0.33 |
ENST00000186436.5 |
TMEM131 |
transmembrane protein 131 |
| chr2_+_148778570 | 0.33 |
ENST00000407073.1 |
MBD5 |
methyl-CpG binding domain protein 5 |
| chr19_+_49617581 | 0.31 |
ENST00000391864.3 |
LIN7B |
lin-7 homolog B (C. elegans) |
| chr12_+_79258444 | 0.30 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr7_-_100493744 | 0.29 |
ENST00000428317.1 ENST00000441605.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr8_+_32406179 | 0.28 |
ENST00000405005.3 |
NRG1 |
neuregulin 1 |
| chr8_+_32405728 | 0.28 |
ENST00000523079.1 ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1 |
neuregulin 1 |
| chr12_+_79258547 | 0.27 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr1_+_114522049 | 0.26 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr1_+_246887349 | 0.26 |
ENST00000366510.3 |
SCCPDH |
saccharopine dehydrogenase (putative) |
| chr11_+_92702886 | 0.26 |
ENST00000257068.2 ENST00000528076.1 |
MTNR1B |
melatonin receptor 1B |
| chr20_+_37353084 | 0.25 |
ENST00000217420.1 |
SLC32A1 |
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr17_+_72772621 | 0.25 |
ENST00000335464.5 ENST00000417024.2 ENST00000578764.1 ENST00000582773.1 ENST00000582330.1 |
TMEM104 |
transmembrane protein 104 |
| chr19_+_49617609 | 0.25 |
ENST00000221459.2 ENST00000486217.2 |
LIN7B |
lin-7 homolog B (C. elegans) |
| chr11_-_65640198 | 0.25 |
ENST00000528176.1 |
EFEMP2 |
EGF containing fibulin-like extracellular matrix protein 2 |
| chr8_-_57123815 | 0.23 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr17_-_1619535 | 0.23 |
ENST00000573075.1 ENST00000574306.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chr19_-_41196534 | 0.23 |
ENST00000252891.4 |
NUMBL |
numb homolog (Drosophila)-like |
| chr1_+_183605200 | 0.23 |
ENST00000304685.4 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr4_-_82393052 | 0.22 |
ENST00000335927.7 ENST00000504863.1 ENST00000264400.2 |
RASGEF1B |
RasGEF domain family, member 1B |
| chr8_+_32405785 | 0.22 |
ENST00000287842.3 |
NRG1 |
neuregulin 1 |
| chr17_-_1619491 | 0.21 |
ENST00000570416.1 ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chr9_+_2621798 | 0.21 |
ENST00000382100.3 |
VLDLR |
very low density lipoprotein receptor |
| chr5_+_40679584 | 0.21 |
ENST00000302472.3 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
| chr5_-_178054014 | 0.21 |
ENST00000520957.1 |
CLK4 |
CDC-like kinase 4 |
| chr8_+_57124245 | 0.21 |
ENST00000521831.1 ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7 |
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr4_-_36246060 | 0.21 |
ENST00000303965.4 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr10_-_13043697 | 0.21 |
ENST00000378825.3 |
CCDC3 |
coiled-coil domain containing 3 |
| chr19_-_54693521 | 0.21 |
ENST00000391754.1 ENST00000245615.1 ENST00000431666.2 |
MBOAT7 |
membrane bound O-acyltransferase domain containing 7 |
| chr6_+_47624172 | 0.20 |
ENST00000507065.1 ENST00000296862.1 |
GPR111 |
G protein-coupled receptor 111 |
| chr2_-_38830030 | 0.20 |
ENST00000410076.1 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
| chr19_-_41196458 | 0.20 |
ENST00000598779.1 |
NUMBL |
numb homolog (Drosophila)-like |
| chr3_+_149689066 | 0.20 |
ENST00000593416.1 |
AC117395.1 |
LOC646903 protein; Uncharacterized protein |
| chr19_-_51472031 | 0.20 |
ENST00000391808.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr20_-_3996036 | 0.19 |
ENST00000336095.6 |
RNF24 |
ring finger protein 24 |
| chr4_-_175443943 | 0.19 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr19_-_15311713 | 0.19 |
ENST00000601011.1 ENST00000263388.2 |
NOTCH3 |
notch 3 |
| chr3_+_134514093 | 0.19 |
ENST00000398015.3 |
EPHB1 |
EPH receptor B1 |
| chr7_-_127032363 | 0.19 |
ENST00000393312.1 |
ZNF800 |
zinc finger protein 800 |
| chr5_+_67584174 | 0.18 |
ENST00000320694.8 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chrX_-_47479246 | 0.18 |
ENST00000295987.7 ENST00000340666.4 |
SYN1 |
synapsin I |
| chr19_-_51472222 | 0.18 |
ENST00000376851.3 |
KLK6 |
kallikrein-related peptidase 6 |
| chr1_+_9648921 | 0.18 |
ENST00000377376.4 ENST00000340305.5 ENST00000340381.6 |
TMEM201 |
transmembrane protein 201 |
| chr15_+_43809797 | 0.18 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr1_-_33647267 | 0.17 |
ENST00000291416.5 |
TRIM62 |
tripartite motif containing 62 |
| chr16_-_850723 | 0.17 |
ENST00000248150.4 |
GNG13 |
guanine nucleotide binding protein (G protein), gamma 13 |
| chr11_+_64009072 | 0.17 |
ENST00000535135.1 ENST00000394540.3 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr3_+_3168692 | 0.17 |
ENST00000402675.1 ENST00000413000.1 |
TRNT1 |
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr17_-_79008373 | 0.17 |
ENST00000577066.1 ENST00000573167.1 |
BAIAP2-AS1 |
BAIAP2 antisense RNA 1 (head to head) |
| chrX_+_133930798 | 0.16 |
ENST00000414371.2 |
FAM122C |
family with sequence similarity 122C |
| chr1_+_159141397 | 0.16 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr9_+_2622085 | 0.16 |
ENST00000382099.2 |
VLDLR |
very low density lipoprotein receptor |
| chr20_-_61569227 | 0.16 |
ENST00000266070.4 ENST00000395335.2 ENST00000266071.5 |
DIDO1 |
death inducer-obliterator 1 |
| chr2_+_202899310 | 0.16 |
ENST00000286201.1 |
FZD7 |
frizzled family receptor 7 |
| chrX_-_112084043 | 0.16 |
ENST00000304758.1 |
AMOT |
angiomotin |
| chrX_+_117957741 | 0.16 |
ENST00000310164.2 |
ZCCHC12 |
zinc finger, CCHC domain containing 12 |
| chr8_-_110704014 | 0.16 |
ENST00000529190.1 ENST00000422135.1 ENST00000419099.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr8_+_38644715 | 0.16 |
ENST00000317827.4 ENST00000379931.3 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr9_-_131709858 | 0.16 |
ENST00000372586.3 |
DOLK |
dolichol kinase |
| chr3_+_101546827 | 0.15 |
ENST00000461724.1 ENST00000483180.1 ENST00000394054.2 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr11_+_46316677 | 0.15 |
ENST00000534787.1 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr10_-_27444143 | 0.15 |
ENST00000477432.1 |
YME1L1 |
YME1-like 1 ATPase |
| chr7_-_108096765 | 0.15 |
ENST00000379024.4 ENST00000351718.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr4_-_119274121 | 0.15 |
ENST00000296498.3 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
| chr20_+_34129770 | 0.15 |
ENST00000348547.2 ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3 |
ERGIC and golgi 3 |
| chr10_-_129924611 | 0.15 |
ENST00000368654.3 |
MKI67 |
marker of proliferation Ki-67 |
| chr15_-_71146460 | 0.15 |
ENST00000344870.4 |
LARP6 |
La ribonucleoprotein domain family, member 6 |
| chr19_+_39786962 | 0.15 |
ENST00000333625.2 |
IFNL1 |
interferon, lambda 1 |
| chr8_+_32406137 | 0.15 |
ENST00000521670.1 |
NRG1 |
neuregulin 1 |
| chr3_-_136471204 | 0.14 |
ENST00000480733.1 ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1 |
stromal antigen 1 |
| chr9_+_37650945 | 0.14 |
ENST00000377765.3 |
FRMPD1 |
FERM and PDZ domain containing 1 |
| chr2_-_220083671 | 0.14 |
ENST00000439002.2 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr2_-_220083692 | 0.14 |
ENST00000265316.3 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr8_-_110703819 | 0.14 |
ENST00000532779.1 ENST00000534578.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr1_-_160040038 | 0.14 |
ENST00000368089.3 |
KCNJ10 |
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr5_+_149569520 | 0.14 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr8_-_27469196 | 0.14 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr12_-_42983478 | 0.14 |
ENST00000345127.3 ENST00000547113.1 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
| chr7_-_108096822 | 0.14 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr1_+_51434357 | 0.14 |
ENST00000396148.1 |
CDKN2C |
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr4_-_175443788 | 0.14 |
ENST00000541923.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr2_-_219433014 | 0.14 |
ENST00000418019.1 ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37 |
ubiquitin specific peptidase 37 |
| chr4_-_175443484 | 0.14 |
ENST00000514584.1 ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr9_-_130517522 | 0.13 |
ENST00000373274.3 ENST00000420366.1 |
SH2D3C |
SH2 domain containing 3C |
| chr8_+_145691411 | 0.13 |
ENST00000301332.2 |
KIFC2 |
kinesin family member C2 |
| chr19_+_44037334 | 0.13 |
ENST00000314228.5 |
ZNF575 |
zinc finger protein 575 |
| chr2_+_61108771 | 0.13 |
ENST00000394479.3 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr12_+_49372251 | 0.13 |
ENST00000293549.3 |
WNT1 |
wingless-type MMTV integration site family, member 1 |
| chr16_+_29690358 | 0.13 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr5_-_9546180 | 0.13 |
ENST00000382496.5 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr2_-_220083076 | 0.13 |
ENST00000295750.4 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr9_-_33264557 | 0.13 |
ENST00000473781.1 ENST00000488499.1 |
BAG1 |
BCL2-associated athanogene |
| chr17_+_30593195 | 0.13 |
ENST00000431505.2 ENST00000269051.4 ENST00000538145.1 |
RHBDL3 |
rhomboid, veinlet-like 3 (Drosophila) |
| chr15_-_83316254 | 0.13 |
ENST00000567678.1 ENST00000450751.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr7_+_100770328 | 0.13 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr3_+_197518100 | 0.13 |
ENST00000438796.2 ENST00000414675.2 ENST00000441090.2 ENST00000334859.4 ENST00000425562.2 |
LRCH3 |
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr19_+_18111927 | 0.13 |
ENST00000379656.3 |
ARRDC2 |
arrestin domain containing 2 |
| chr16_+_2588012 | 0.12 |
ENST00000354836.5 ENST00000389224.3 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr15_+_91446157 | 0.12 |
ENST00000559717.1 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
| chr5_-_141030943 | 0.12 |
ENST00000522783.1 ENST00000519800.1 ENST00000435817.2 |
FCHSD1 |
FCH and double SH3 domains 1 |
| chr11_+_43964055 | 0.12 |
ENST00000528572.1 |
C11orf96 |
chromosome 11 open reading frame 96 |
| chr19_-_36523709 | 0.12 |
ENST00000592017.1 ENST00000360535.4 |
CLIP3 |
CAP-GLY domain containing linker protein 3 |
| chr11_+_20044600 | 0.12 |
ENST00000311043.8 |
NAV2 |
neuron navigator 2 |
| chr2_-_158732340 | 0.12 |
ENST00000539637.1 ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1 |
activin A receptor, type I |
| chr15_-_41408339 | 0.12 |
ENST00000401393.3 |
INO80 |
INO80 complex subunit |
| chr10_-_62761188 | 0.12 |
ENST00000357917.4 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr19_+_35168633 | 0.12 |
ENST00000505365.2 |
ZNF302 |
zinc finger protein 302 |
| chrX_+_153686614 | 0.12 |
ENST00000369682.3 |
PLXNA3 |
plexin A3 |
| chr10_+_105036909 | 0.12 |
ENST00000369849.4 |
INA |
internexin neuronal intermediate filament protein, alpha |
| chr1_-_37499726 | 0.12 |
ENST00000373091.3 ENST00000373093.4 |
GRIK3 |
glutamate receptor, ionotropic, kainate 3 |
| chrX_-_19905703 | 0.12 |
ENST00000397821.3 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr10_+_131265443 | 0.12 |
ENST00000306010.7 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
| chr21_-_28338732 | 0.12 |
ENST00000284987.5 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chr2_+_69002052 | 0.12 |
ENST00000497079.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr5_-_172662303 | 0.12 |
ENST00000517440.1 ENST00000329198.4 |
NKX2-5 |
NK2 homeobox 5 |
| chr6_+_31865552 | 0.12 |
ENST00000469372.1 ENST00000497706.1 |
C2 |
complement component 2 |
| chr16_+_2587965 | 0.12 |
ENST00000342085.4 ENST00000566659.1 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr2_-_97405775 | 0.11 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr7_+_95401851 | 0.11 |
ENST00000447467.2 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
| chr16_+_53164833 | 0.11 |
ENST00000564845.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr16_+_2198604 | 0.11 |
ENST00000210187.6 |
RAB26 |
RAB26, member RAS oncogene family |
| chr19_+_55795493 | 0.11 |
ENST00000309383.1 |
BRSK1 |
BR serine/threonine kinase 1 |
| chr9_-_113800317 | 0.11 |
ENST00000374431.3 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr11_+_66059339 | 0.11 |
ENST00000327259.4 |
TMEM151A |
transmembrane protein 151A |
| chr16_-_2185899 | 0.11 |
ENST00000262304.4 ENST00000423118.1 |
PKD1 |
polycystic kidney disease 1 (autosomal dominant) |
| chr15_+_76629064 | 0.11 |
ENST00000290759.4 |
ISL2 |
ISL LIM homeobox 2 |
| chr1_+_65886262 | 0.11 |
ENST00000371065.4 |
LEPROT |
leptin receptor overlapping transcript |
| chr15_-_75744014 | 0.11 |
ENST00000394947.3 ENST00000565264.1 |
SIN3A |
SIN3 transcription regulator family member A |
| chr1_-_38512450 | 0.11 |
ENST00000373012.2 |
POU3F1 |
POU class 3 homeobox 1 |
| chr12_-_57634475 | 0.11 |
ENST00000393825.1 |
NDUFA4L2 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr15_-_93632421 | 0.11 |
ENST00000329082.7 |
RGMA |
repulsive guidance molecule family member a |
| chr1_+_107683436 | 0.11 |
ENST00000370068.1 |
NTNG1 |
netrin G1 |
| chr11_-_65548265 | 0.11 |
ENST00000532090.2 |
AP5B1 |
adaptor-related protein complex 5, beta 1 subunit |
| chr3_-_169487617 | 0.11 |
ENST00000330368.2 |
ACTRT3 |
actin-related protein T3 |
| chr15_-_75743991 | 0.11 |
ENST00000567289.1 |
SIN3A |
SIN3 transcription regulator family member A |
| chr16_-_2264779 | 0.11 |
ENST00000333503.7 |
PGP |
phosphoglycolate phosphatase |
| chr19_-_40724246 | 0.11 |
ENST00000311308.6 |
TTC9B |
tetratricopeptide repeat domain 9B |
| chr17_+_48796905 | 0.11 |
ENST00000505658.1 ENST00000393227.2 ENST00000240304.1 ENST00000311571.3 ENST00000505619.1 ENST00000544170.1 ENST00000510984.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr4_-_83351294 | 0.11 |
ENST00000502762.1 |
HNRNPDL |
heterogeneous nuclear ribonucleoprotein D-like |
| chr5_-_33892046 | 0.11 |
ENST00000352040.3 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr3_-_9994021 | 0.11 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chr15_+_32322685 | 0.11 |
ENST00000454250.3 ENST00000306901.3 |
CHRNA7 |
cholinergic receptor, nicotinic, alpha 7 (neuronal) |
| chr6_-_137113604 | 0.10 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr10_-_64028466 | 0.10 |
ENST00000395265.1 ENST00000373789.3 ENST00000395260.3 |
RTKN2 |
rhotekin 2 |
| chr17_+_59477233 | 0.10 |
ENST00000240328.3 |
TBX2 |
T-box 2 |
| chr4_-_57976544 | 0.10 |
ENST00000295666.4 ENST00000537922.1 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
| chr16_+_2587998 | 0.10 |
ENST00000441549.3 ENST00000268673.7 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr17_-_62502399 | 0.10 |
ENST00000450599.2 ENST00000585060.1 |
DDX5 |
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr22_+_37956453 | 0.10 |
ENST00000249014.4 |
CDC42EP1 |
CDC42 effector protein (Rho GTPase binding) 1 |
| chr11_+_117198801 | 0.10 |
ENST00000527609.1 ENST00000533570.1 |
CEP164 |
centrosomal protein 164kDa |
| chr16_+_69140122 | 0.10 |
ENST00000219322.3 |
HAS3 |
hyaluronan synthase 3 |
| chr19_+_35168567 | 0.10 |
ENST00000457781.2 ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302 |
zinc finger protein 302 |
| chr9_-_132515302 | 0.10 |
ENST00000340607.4 |
PTGES |
prostaglandin E synthase |
| chr20_+_53092123 | 0.10 |
ENST00000262593.5 |
DOK5 |
docking protein 5 |
| chr2_+_121103706 | 0.10 |
ENST00000295228.3 |
INHBB |
inhibin, beta B |
| chr5_+_67511524 | 0.10 |
ENST00000521381.1 ENST00000521657.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr15_+_32322709 | 0.10 |
ENST00000455693.2 |
CHRNA7 |
cholinergic receptor, nicotinic, alpha 7 (neuronal) |
| chr5_-_178054105 | 0.10 |
ENST00000316308.4 |
CLK4 |
CDC-like kinase 4 |
| chr20_+_34894247 | 0.10 |
ENST00000373913.3 |
DLGAP4 |
discs, large (Drosophila) homolog-associated protein 4 |
| chr3_-_88108212 | 0.10 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr6_+_30524663 | 0.10 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr15_-_27184664 | 0.10 |
ENST00000541819.2 |
GABRB3 |
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr22_+_45898712 | 0.10 |
ENST00000455233.1 ENST00000348697.2 ENST00000402984.3 ENST00000262722.7 ENST00000327858.6 ENST00000442170.2 ENST00000340923.5 ENST00000439835.1 |
FBLN1 |
fibulin 1 |
| chr3_-_87040233 | 0.10 |
ENST00000398399.2 |
VGLL3 |
vestigial like 3 (Drosophila) |
| chr8_-_27468945 | 0.10 |
ENST00000405140.3 |
CLU |
clusterin |
| chr19_-_16682987 | 0.10 |
ENST00000431408.1 ENST00000436553.2 ENST00000595753.1 |
SLC35E1 |
solute carrier family 35, member E1 |
| chr13_-_110959478 | 0.10 |
ENST00000543140.1 ENST00000375820.4 |
COL4A1 |
collagen, type IV, alpha 1 |
| chr17_-_62502022 | 0.10 |
ENST00000578804.1 |
DDX5 |
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr3_-_124774802 | 0.10 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr14_-_74417096 | 0.10 |
ENST00000286544.3 |
FAM161B |
family with sequence similarity 161, member B |
| chr1_-_160001737 | 0.10 |
ENST00000368090.2 |
PIGM |
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr2_+_191745535 | 0.10 |
ENST00000320717.3 |
GLS |
glutaminase |
| chr1_+_236305826 | 0.09 |
ENST00000366592.3 ENST00000366591.4 |
GPR137B |
G protein-coupled receptor 137B |
| chrX_-_108868390 | 0.09 |
ENST00000372101.2 |
KCNE1L |
KCNE1-like |
| chr5_-_14871866 | 0.09 |
ENST00000284268.6 |
ANKH |
ANKH inorganic pyrophosphate transport regulator |
| chr1_+_178062855 | 0.09 |
ENST00000448150.3 |
RASAL2 |
RAS protein activator like 2 |
| chr17_+_57697216 | 0.09 |
ENST00000393043.1 ENST00000269122.3 |
CLTC |
clathrin, heavy chain (Hc) |
| chr15_+_41952591 | 0.09 |
ENST00000566718.1 ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA |
MGA, MAX dimerization protein |
| chr6_-_111804905 | 0.09 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chrX_-_135056106 | 0.09 |
ENST00000433339.2 |
MMGT1 |
membrane magnesium transporter 1 |
| chr5_+_109025067 | 0.09 |
ENST00000261483.4 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
| chr17_-_62502639 | 0.09 |
ENST00000225792.5 ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5 |
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr14_+_23352374 | 0.09 |
ENST00000267396.4 ENST00000536884.1 |
REM2 |
RAS (RAD and GEM)-like GTP binding 2 |
| chr14_-_75079026 | 0.09 |
ENST00000261978.4 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.2 | 0.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 0.5 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.4 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.4 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.9 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.5 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.2 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 0.2 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.1 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.1 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.1 | 0.4 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.4 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 0.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.2 | GO:1902725 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 1.0 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.3 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.5 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 0.0 | 0.1 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.2 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.2 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) septum secundum development(GO:0003285) |
| 0.0 | 0.5 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.4 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0072237 | distal tubule morphogenesis(GO:0072156) metanephric proximal tubule development(GO:0072237) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.2 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.2 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.8 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.4 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.2 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.1 | GO:1901895 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0015880 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.8 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.0 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.0 | 0.2 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.1 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.1 | GO:0061086 | regulation of mRNA export from nucleus(GO:0010793) negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.0 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 1.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.0 | GO:0030849 | Y chromosome(GO:0000806) autosome(GO:0030849) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 0.4 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.5 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.8 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.3 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.3 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.5 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0008523 | L-ascorbate:sodium symporter activity(GO:0008520) sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.3 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.0 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0042954 | apolipoprotein receptor activity(GO:0030226) lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0052848 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.0 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |