Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
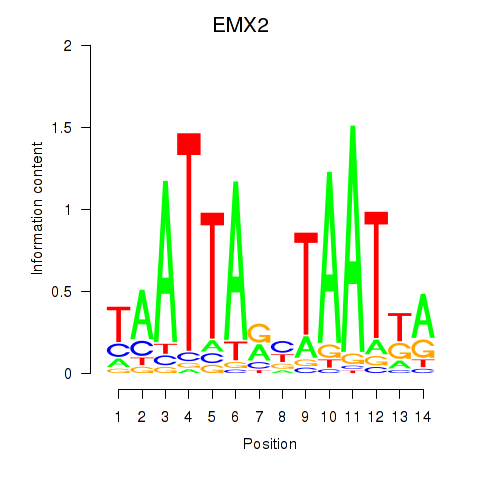
Results for EMX2
Z-value: 0.70
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.10 | EMX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg19_v2_chr10_+_119301928_119301955 | 0.35 | 4.0e-01 | Click! |
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_72926145 | 1.17 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr11_-_63376013 | 1.15 |
ENST00000540943.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chr12_-_91574142 | 1.03 |
ENST00000547937.1 |
DCN |
decorin |
| chr5_-_111093081 | 0.98 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093167 | 0.86 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr5_-_111312622 | 0.79 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr5_+_32788945 | 0.72 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr5_-_75919253 | 0.65 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr4_-_138453606 | 0.60 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr5_-_111092930 | 0.58 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr17_+_1674982 | 0.55 |
ENST00000572048.1 ENST00000573763.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr11_+_5710919 | 0.53 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr8_-_13134045 | 0.49 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr21_-_31869451 | 0.48 |
ENST00000334058.2 |
KRTAP19-4 |
keratin associated protein 19-4 |
| chr19_+_50016610 | 0.47 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr5_-_75919217 | 0.46 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr6_+_72922505 | 0.42 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr1_+_78383813 | 0.40 |
ENST00000342754.5 |
NEXN |
nexilin (F actin binding protein) |
| chr2_-_190044480 | 0.40 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr10_-_79398250 | 0.39 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_-_179343226 | 0.38 |
ENST00000434643.2 |
FKBP7 |
FK506 binding protein 7 |
| chr12_+_60083118 | 0.38 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr6_+_72922590 | 0.37 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr10_-_79398127 | 0.36 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_+_78470530 | 0.36 |
ENST00000370763.5 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr7_+_138915102 | 0.35 |
ENST00000486663.1 |
UBN2 |
ubinuclein 2 |
| chr10_-_79397740 | 0.35 |
ENST00000372440.1 ENST00000480683.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr8_+_70404996 | 0.34 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr9_-_16728161 | 0.32 |
ENST00000603713.1 ENST00000603313.1 |
BNC2 |
basonuclin 2 |
| chr2_-_99279928 | 0.32 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr14_+_52164820 | 0.32 |
ENST00000554167.1 |
FRMD6 |
FERM domain containing 6 |
| chr9_-_99540328 | 0.32 |
ENST00000223428.4 ENST00000375231.1 ENST00000374641.3 |
ZNF510 |
zinc finger protein 510 |
| chr1_+_192778161 | 0.32 |
ENST00000235382.5 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
| chr2_-_207082748 | 0.31 |
ENST00000407325.2 ENST00000411719.1 |
GPR1 |
G protein-coupled receptor 1 |
| chr1_+_79115503 | 0.31 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr7_+_90338712 | 0.30 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr4_-_138453559 | 0.29 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr4_+_156824840 | 0.28 |
ENST00000536354.2 |
TDO2 |
tryptophan 2,3-dioxygenase |
| chr17_-_67138015 | 0.27 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr14_+_74034310 | 0.26 |
ENST00000538782.1 |
ACOT2 |
acyl-CoA thioesterase 2 |
| chr1_-_85870177 | 0.26 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr15_-_20193370 | 0.25 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr19_-_44388116 | 0.24 |
ENST00000587539.1 |
ZNF404 |
zinc finger protein 404 |
| chrX_+_77166172 | 0.24 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr12_+_59989918 | 0.23 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_+_66276550 | 0.22 |
ENST00000419755.3 |
CTD-3074O7.11 |
Bardet-Biedl syndrome 1 protein |
| chrX_+_55744166 | 0.22 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chrX_+_120181457 | 0.21 |
ENST00000328078.1 |
GLUD2 |
glutamate dehydrogenase 2 |
| chr15_-_57025759 | 0.21 |
ENST00000267807.7 |
ZNF280D |
zinc finger protein 280D |
| chr10_+_15074190 | 0.21 |
ENST00000428897.1 ENST00000413672.1 |
OLAH |
oleoyl-ACP hydrolase |
| chr4_+_74347400 | 0.21 |
ENST00000226355.3 |
AFM |
afamin |
| chr14_-_47351391 | 0.20 |
ENST00000399222.3 |
MDGA2 |
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr7_-_44887620 | 0.20 |
ENST00000349299.3 ENST00000521529.1 ENST00000308153.4 ENST00000350771.3 ENST00000222690.6 ENST00000381124.5 ENST00000437072.1 ENST00000446531.1 |
H2AFV |
H2A histone family, member V |
| chrX_+_55744228 | 0.20 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr5_+_81575281 | 0.20 |
ENST00000380167.4 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr7_-_140482926 | 0.19 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr2_+_161993465 | 0.19 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr4_+_174818390 | 0.19 |
ENST00000509968.1 ENST00000512263.1 |
RP11-161D15.1 |
RP11-161D15.1 |
| chr3_-_49466686 | 0.19 |
ENST00000273598.3 ENST00000436744.2 |
NICN1 |
nicolin 1 |
| chr1_-_110283660 | 0.19 |
ENST00000361066.2 |
GSTM3 |
glutathione S-transferase mu 3 (brain) |
| chr17_-_295730 | 0.19 |
ENST00000329099.4 |
FAM101B |
family with sequence similarity 101, member B |
| chr12_+_103981044 | 0.19 |
ENST00000388887.2 |
STAB2 |
stabilin 2 |
| chr6_-_29343068 | 0.18 |
ENST00000396806.3 |
OR12D3 |
olfactory receptor, family 12, subfamily D, member 3 |
| chr4_+_56212505 | 0.18 |
ENST00000505210.1 |
SRD5A3 |
steroid 5 alpha-reductase 3 |
| chr2_-_85625857 | 0.18 |
ENST00000453973.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr6_-_26235206 | 0.18 |
ENST00000244534.5 |
HIST1H1D |
histone cluster 1, H1d |
| chr10_+_1095416 | 0.18 |
ENST00000358220.1 |
WDR37 |
WD repeat domain 37 |
| chr6_+_101846664 | 0.18 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr5_-_76788317 | 0.18 |
ENST00000296679.4 |
WDR41 |
WD repeat domain 41 |
| chr1_+_174844645 | 0.18 |
ENST00000486220.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr8_-_93029865 | 0.17 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr16_+_446713 | 0.17 |
ENST00000397722.1 ENST00000454619.1 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
| chr4_+_56212270 | 0.17 |
ENST00000264228.4 |
SRD5A3 |
steroid 5 alpha-reductase 3 |
| chr10_+_91092241 | 0.17 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr11_+_12115543 | 0.17 |
ENST00000537344.1 ENST00000532179.1 ENST00000526065.1 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr19_-_58864848 | 0.17 |
ENST00000263100.3 |
A1BG |
alpha-1-B glycoprotein |
| chr17_+_39240459 | 0.16 |
ENST00000391417.4 |
KRTAP4-7 |
keratin associated protein 4-7 |
| chr3_-_127455200 | 0.16 |
ENST00000398101.3 |
MGLL |
monoglyceride lipase |
| chr11_-_124981475 | 0.16 |
ENST00000532156.1 ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218 |
transmembrane protein 218 |
| chr20_+_15177480 | 0.16 |
ENST00000402914.1 |
MACROD2 |
MACRO domain containing 2 |
| chr3_+_130745688 | 0.15 |
ENST00000510769.1 ENST00000429253.2 ENST00000356918.4 ENST00000510688.1 ENST00000511262.1 ENST00000383366.4 |
NEK11 |
NIMA-related kinase 11 |
| chr2_+_161993412 | 0.15 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr1_+_84609944 | 0.15 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr19_-_35264089 | 0.15 |
ENST00000588760.1 ENST00000329285.8 ENST00000587354.2 |
ZNF599 |
zinc finger protein 599 |
| chr9_+_125133315 | 0.15 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_+_178977546 | 0.15 |
ENST00000319449.4 ENST00000377001.2 |
RUFY1 |
RUN and FYVE domain containing 1 |
| chr16_+_69599861 | 0.14 |
ENST00000354436.2 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr6_+_28092338 | 0.14 |
ENST00000340487.4 |
ZSCAN16 |
zinc finger and SCAN domain containing 16 |
| chr12_+_26164645 | 0.14 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr19_-_37697976 | 0.14 |
ENST00000588873.1 |
CTC-454I21.3 |
Uncharacterized protein; Zinc finger protein 585B |
| chr6_+_63921399 | 0.14 |
ENST00000356170.3 |
FKBP1C |
FK506 binding protein 1C |
| chr20_+_57414743 | 0.14 |
ENST00000313949.7 |
GNAS |
GNAS complex locus |
| chr16_-_28937027 | 0.14 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr17_-_34503867 | 0.14 |
ENST00000589443.1 ENST00000454519.3 ENST00000586369.1 |
TBC1D3B |
TBC1 domain family, member 3B |
| chr17_+_66521936 | 0.14 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr12_-_71533055 | 0.14 |
ENST00000552128.1 |
TSPAN8 |
tetraspanin 8 |
| chr8_-_57123815 | 0.14 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr8_-_133687778 | 0.13 |
ENST00000518642.1 |
LRRC6 |
leucine rich repeat containing 6 |
| chr14_-_105708942 | 0.13 |
ENST00000549655.1 |
BRF1 |
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr3_-_186288097 | 0.13 |
ENST00000446782.1 |
TBCCD1 |
TBCC domain containing 1 |
| chr1_+_244998918 | 0.13 |
ENST00000366528.3 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
| chr5_-_88119580 | 0.13 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr11_-_104916034 | 0.13 |
ENST00000528513.1 ENST00000375706.2 ENST00000375704.3 |
CARD16 |
caspase recruitment domain family, member 16 |
| chr17_-_15244894 | 0.13 |
ENST00000338696.2 ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3 |
tektin 3 |
| chr5_+_54455946 | 0.13 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr8_-_133687813 | 0.13 |
ENST00000250173.1 ENST00000519595.1 |
LRRC6 |
leucine rich repeat containing 6 |
| chr12_-_53171128 | 0.13 |
ENST00000332411.2 |
KRT76 |
keratin 76 |
| chr11_-_26593649 | 0.12 |
ENST00000455601.2 |
MUC15 |
mucin 15, cell surface associated |
| chr17_-_34757039 | 0.12 |
ENST00000455054.2 ENST00000308078.7 |
TBC1D3H TBC1D3C |
TBC1 domain family, member 3H TBC1 domain family, member 3C |
| chr3_+_40518599 | 0.12 |
ENST00000314686.5 ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619 |
zinc finger protein 619 |
| chr13_-_107214291 | 0.12 |
ENST00000375926.1 |
ARGLU1 |
arginine and glutamate rich 1 |
| chr5_+_118812237 | 0.12 |
ENST00000513628.1 |
HSD17B4 |
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr14_-_80697396 | 0.12 |
ENST00000557010.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr12_+_94071341 | 0.12 |
ENST00000542893.2 |
CRADD |
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr7_+_138145076 | 0.11 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr8_-_25281747 | 0.11 |
ENST00000421054.2 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr5_+_161495038 | 0.11 |
ENST00000393933.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr6_+_27925019 | 0.11 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr1_-_54405773 | 0.11 |
ENST00000371376.1 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr11_-_26593677 | 0.11 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr1_+_10292308 | 0.11 |
ENST00000377081.1 |
KIF1B |
kinesin family member 1B |
| chr11_-_327537 | 0.11 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr1_-_19615744 | 0.11 |
ENST00000361640.4 |
AKR7A3 |
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr5_+_118812294 | 0.10 |
ENST00000509514.1 |
HSD17B4 |
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr15_-_99789736 | 0.10 |
ENST00000560235.1 ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr19_-_14945933 | 0.10 |
ENST00000322301.3 |
OR7A5 |
olfactory receptor, family 7, subfamily A, member 5 |
| chr11_-_26593779 | 0.10 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr3_-_58523010 | 0.10 |
ENST00000459701.2 ENST00000302819.5 |
ACOX2 |
acyl-CoA oxidase 2, branched chain |
| chr5_+_140514782 | 0.10 |
ENST00000231134.5 |
PCDHB5 |
protocadherin beta 5 |
| chr5_+_143191726 | 0.10 |
ENST00000289448.2 |
HMHB1 |
histocompatibility (minor) HB-1 |
| chr12_+_94071129 | 0.10 |
ENST00000552983.1 ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD |
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr20_-_33999766 | 0.10 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr15_+_64680003 | 0.10 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chrX_+_135730297 | 0.10 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chrX_+_36053908 | 0.10 |
ENST00000378660.2 |
CHDC2 |
calponin homology domain containing 2 |
| chr17_+_62461569 | 0.10 |
ENST00000603557.1 ENST00000605096.1 |
MILR1 |
mast cell immunoglobulin-like receptor 1 |
| chr2_-_105030466 | 0.09 |
ENST00000449772.1 |
AC068535.3 |
AC068535.3 |
| chr16_-_46865286 | 0.09 |
ENST00000285697.4 |
C16orf87 |
chromosome 16 open reading frame 87 |
| chr3_+_195413160 | 0.09 |
ENST00000599448.1 |
LINC00969 |
long intergenic non-protein coding RNA 969 |
| chr5_+_147774275 | 0.09 |
ENST00000513826.1 |
FBXO38 |
F-box protein 38 |
| chr11_+_117198545 | 0.09 |
ENST00000533153.1 ENST00000278935.3 ENST00000525416.1 |
CEP164 |
centrosomal protein 164kDa |
| chr19_+_58144529 | 0.09 |
ENST00000347302.3 ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211 |
zinc finger protein 211 |
| chr10_-_27529486 | 0.09 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr4_-_101439148 | 0.09 |
ENST00000511970.1 ENST00000502569.1 ENST00000305864.3 |
EMCN |
endomucin |
| chr2_+_105050794 | 0.09 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr10_-_99771079 | 0.09 |
ENST00000309155.3 |
CRTAC1 |
cartilage acidic protein 1 |
| chr7_+_13141097 | 0.09 |
ENST00000411542.1 |
AC011288.2 |
AC011288.2 |
| chr1_+_146373546 | 0.09 |
ENST00000446760.2 |
NBPF12 |
neuroblastoma breakpoint family, member 12 |
| chrX_+_47444613 | 0.09 |
ENST00000445623.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr5_-_114938090 | 0.09 |
ENST00000427199.2 |
TICAM2 |
toll-like receptor adaptor molecule 2 |
| chr15_+_84115868 | 0.09 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr16_-_30102547 | 0.09 |
ENST00000279386.2 |
TBX6 |
T-box 6 |
| chr19_+_15852203 | 0.09 |
ENST00000305892.1 |
OR10H3 |
olfactory receptor, family 10, subfamily H, member 3 |
| chr22_+_24198890 | 0.09 |
ENST00000345044.6 |
SLC2A11 |
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr9_+_75766763 | 0.09 |
ENST00000456643.1 ENST00000415424.1 |
ANXA1 |
annexin A1 |
| chr16_+_28648975 | 0.09 |
ENST00000529716.1 |
NPIPB8 |
nuclear pore complex interacting protein family, member B8 |
| chr6_+_63921351 | 0.08 |
ENST00000370659.1 |
FKBP1C |
FK506 binding protein 1C |
| chr3_+_158787041 | 0.08 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr14_+_24099318 | 0.08 |
ENST00000432832.2 |
DHRS2 |
dehydrogenase/reductase (SDR family) member 2 |
| chrX_+_18443703 | 0.08 |
ENST00000379996.3 |
CDKL5 |
cyclin-dependent kinase-like 5 |
| chrX_+_135730373 | 0.08 |
ENST00000370628.2 |
CD40LG |
CD40 ligand |
| chr4_-_89205705 | 0.08 |
ENST00000295908.7 ENST00000510548.2 ENST00000508256.1 |
PPM1K |
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr12_+_58176525 | 0.08 |
ENST00000543727.1 ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM |
Ts translation elongation factor, mitochondrial |
| chr13_+_108921977 | 0.08 |
ENST00000430559.1 ENST00000375887.4 |
TNFSF13B |
tumor necrosis factor (ligand) superfamily, member 13b |
| chr1_+_197382957 | 0.08 |
ENST00000367397.1 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr6_-_74233480 | 0.08 |
ENST00000455918.1 |
EEF1A1 |
eukaryotic translation elongation factor 1 alpha 1 |
| chr20_-_48732472 | 0.08 |
ENST00000340309.3 ENST00000415862.2 ENST00000371677.3 ENST00000420027.2 |
UBE2V1 |
ubiquitin-conjugating enzyme E2 variant 1 |
| chr2_+_122494676 | 0.08 |
ENST00000455432.1 |
TSN |
translin |
| chr19_-_51920952 | 0.08 |
ENST00000356298.5 ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10 |
sialic acid binding Ig-like lectin 10 |
| chr17_+_5031687 | 0.07 |
ENST00000250066.6 ENST00000304328.5 |
USP6 |
ubiquitin specific peptidase 6 (Tre-2 oncogene) |
| chr3_-_48956818 | 0.07 |
ENST00000408959.2 |
ARIH2OS |
ariadne homolog 2 opposite strand |
| chr9_-_21217310 | 0.07 |
ENST00000380216.1 |
IFNA16 |
interferon, alpha 16 |
| chr12_-_11091862 | 0.07 |
ENST00000537503.1 |
TAS2R14 |
taste receptor, type 2, member 14 |
| chr5_+_112312416 | 0.07 |
ENST00000389063.2 |
DCP2 |
decapping mRNA 2 |
| chr21_+_35552978 | 0.07 |
ENST00000428914.2 ENST00000609062.1 ENST00000609947.1 |
LINC00310 |
long intergenic non-protein coding RNA 310 |
| chr9_-_21202204 | 0.07 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr1_-_146082633 | 0.07 |
ENST00000605317.1 ENST00000604938.1 ENST00000339388.5 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr3_-_155011483 | 0.07 |
ENST00000489090.1 |
RP11-451G4.2 |
RP11-451G4.2 |
| chr16_+_28763108 | 0.07 |
ENST00000357796.3 ENST00000550983.1 |
NPIPB9 |
nuclear pore complex interacting protein family, member B9 |
| chr2_+_242254507 | 0.07 |
ENST00000391973.2 |
SEPT2 |
septin 2 |
| chr21_+_31768348 | 0.07 |
ENST00000355459.2 |
KRTAP13-1 |
keratin associated protein 13-1 |
| chr6_+_29079668 | 0.07 |
ENST00000377169.1 |
OR2J3 |
olfactory receptor, family 2, subfamily J, member 3 |
| chr3_+_111393501 | 0.07 |
ENST00000393934.3 |
PLCXD2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr2_+_11682790 | 0.07 |
ENST00000389825.3 ENST00000381483.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr13_+_103459704 | 0.07 |
ENST00000602836.1 |
BIVM-ERCC5 |
BIVM-ERCC5 readthrough |
| chr10_+_51187938 | 0.06 |
ENST00000311663.5 |
FAM21D |
family with sequence similarity 21, member D |
| chr3_-_130745571 | 0.06 |
ENST00000514044.1 ENST00000264992.3 |
ASTE1 |
asteroid homolog 1 (Drosophila) |
| chr6_+_28317685 | 0.06 |
ENST00000252211.2 ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3 |
zinc finger with KRAB and SCAN domains 3 |
| chr16_-_21436459 | 0.06 |
ENST00000448012.2 ENST00000504841.2 ENST00000419180.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr2_-_197226875 | 0.06 |
ENST00000409111.1 |
HECW2 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr4_+_113568207 | 0.06 |
ENST00000511529.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr12_-_53715328 | 0.06 |
ENST00000547757.1 ENST00000394384.3 ENST00000209873.4 |
AAAS |
achalasia, adrenocortical insufficiency, alacrimia |
| chr4_-_113437328 | 0.06 |
ENST00000313341.3 |
NEUROG2 |
neurogenin 2 |
| chr16_-_28374829 | 0.06 |
ENST00000532254.1 |
NPIPB6 |
nuclear pore complex interacting protein family, member B6 |
| chr6_-_74231444 | 0.06 |
ENST00000331523.2 ENST00000356303.2 |
EEF1A1 |
eukaryotic translation elongation factor 1 alpha 1 |
| chr5_-_86534822 | 0.06 |
ENST00000445770.2 |
AC008394.1 |
Uncharacterized protein |
| chr17_-_4167142 | 0.06 |
ENST00000570535.1 ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chr6_+_42018614 | 0.06 |
ENST00000465926.1 ENST00000482432.1 |
TAF8 |
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr10_-_47239738 | 0.06 |
ENST00000413193.2 |
AGAP10 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr18_+_6729725 | 0.06 |
ENST00000400091.2 ENST00000583410.1 ENST00000584387.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.1 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 1.7 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.6 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.5 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 1.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.3 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.0 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.0 | 0.1 | GO:0009304 | tRNA transcription(GO:0009304) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.0 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 1.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.6 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 1.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.4 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.2 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.2 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |