Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for EOMES
Z-value: 0.68
Transcription factors associated with EOMES
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EOMES
|
ENSG00000163508.8 | EOMES |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EOMES | hg19_v2_chr3_-_27763803_27763822 | 0.57 | 1.4e-01 | Click! |
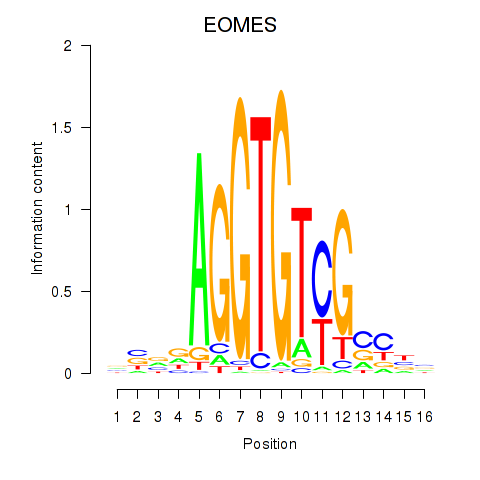
Activity profile of EOMES motif
Sorted Z-values of EOMES motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EOMES
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_28681950 | 0.63 |
ENST00000251081.6 |
DSC2 |
desmocollin 2 |
| chr1_+_70876926 | 0.34 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr1_-_207095324 | 0.34 |
ENST00000530505.1 ENST00000367091.3 ENST00000442471.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr2_-_229046361 | 0.33 |
ENST00000392056.3 |
SPHKAP |
SPHK1 interactor, AKAP domain containing |
| chr13_-_20767037 | 0.33 |
ENST00000382848.4 |
GJB2 |
gap junction protein, beta 2, 26kDa |
| chr14_+_61995722 | 0.33 |
ENST00000556347.1 |
RP11-47I22.4 |
RP11-47I22.4 |
| chr2_-_229046330 | 0.32 |
ENST00000344657.5 |
SPHKAP |
SPHK1 interactor, AKAP domain containing |
| chr1_+_60280458 | 0.32 |
ENST00000455990.1 ENST00000371208.3 |
HOOK1 |
hook microtubule-tethering protein 1 |
| chr1_+_1981890 | 0.31 |
ENST00000378567.3 ENST00000468310.1 |
PRKCZ |
protein kinase C, zeta |
| chr16_-_2031464 | 0.31 |
ENST00000356120.4 ENST00000354249.4 |
NOXO1 |
NADPH oxidase organizer 1 |
| chr15_+_41136586 | 0.31 |
ENST00000431806.1 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr1_+_70876891 | 0.31 |
ENST00000411986.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr17_+_37784749 | 0.30 |
ENST00000394265.1 ENST00000394267.2 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_+_15256230 | 0.30 |
ENST00000376028.4 ENST00000400798.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr1_-_184943610 | 0.28 |
ENST00000367511.3 |
FAM129A |
family with sequence similarity 129, member A |
| chr20_-_18038521 | 0.28 |
ENST00000278780.6 |
OVOL2 |
ovo-like zinc finger 2 |
| chr10_+_95653687 | 0.25 |
ENST00000371408.3 ENST00000427197.1 |
SLC35G1 |
solute carrier family 35, member G1 |
| chr19_-_51456321 | 0.25 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr19_-_51456344 | 0.25 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr15_+_89182178 | 0.24 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr1_-_207095212 | 0.24 |
ENST00000420007.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr1_-_242687989 | 0.24 |
ENST00000442594.2 |
PLD5 |
phospholipase D family, member 5 |
| chr14_+_21510385 | 0.24 |
ENST00000298690.4 |
RNASE7 |
ribonuclease, RNase A family, 7 |
| chr5_+_66124590 | 0.22 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr10_-_10836865 | 0.22 |
ENST00000446372.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr15_+_89182156 | 0.22 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr2_+_95940186 | 0.20 |
ENST00000403131.2 ENST00000317668.4 ENST00000317620.9 |
PROM2 |
prominin 2 |
| chr21_-_47352477 | 0.20 |
ENST00000593412.1 |
PRED62 |
Uncharacterized protein |
| chr1_-_31769656 | 0.20 |
ENST00000446633.2 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr3_+_111805182 | 0.20 |
ENST00000430855.1 ENST00000431717.2 ENST00000264848.5 |
C3orf52 |
chromosome 3 open reading frame 52 |
| chrX_+_135618258 | 0.20 |
ENST00000440515.1 ENST00000456412.1 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr8_-_9760839 | 0.20 |
ENST00000519461.1 ENST00000517675.1 |
LINC00599 |
long intergenic non-protein coding RNA 599 |
| chrX_+_51149767 | 0.19 |
ENST00000342995.2 |
CXorf67 |
chromosome X open reading frame 67 |
| chrX_+_35816908 | 0.19 |
ENST00000399985.1 |
MAGEB16 |
melanoma antigen family B, 16 |
| chr1_+_14925173 | 0.18 |
ENST00000376030.2 ENST00000503743.1 ENST00000422387.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr15_+_89181974 | 0.18 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr18_-_74839891 | 0.18 |
ENST00000581878.1 |
MBP |
myelin basic protein |
| chr17_+_9548845 | 0.18 |
ENST00000570475.1 ENST00000285199.7 |
USP43 |
ubiquitin specific peptidase 43 |
| chr12_+_93965609 | 0.17 |
ENST00000549887.1 ENST00000551556.1 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr3_-_50649192 | 0.17 |
ENST00000443053.2 ENST00000348721.3 |
CISH |
cytokine inducible SH2-containing protein |
| chr4_-_4291861 | 0.17 |
ENST00000343470.4 |
LYAR |
Ly1 antibody reactive |
| chr4_-_4291761 | 0.17 |
ENST00000513174.1 |
LYAR |
Ly1 antibody reactive |
| chr2_+_220492287 | 0.17 |
ENST00000273063.6 ENST00000373762.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr4_-_4291748 | 0.17 |
ENST00000452476.1 |
LYAR |
Ly1 antibody reactive |
| chr1_-_31769595 | 0.16 |
ENST00000263694.4 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr19_+_48867652 | 0.16 |
ENST00000344846.2 |
SYNGR4 |
synaptogyrin 4 |
| chr13_-_102068769 | 0.16 |
ENST00000376196.3 |
NALCN |
sodium leak channel, non-selective |
| chr2_+_220492116 | 0.16 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr2_+_220492373 | 0.16 |
ENST00000317151.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr7_+_94285637 | 0.16 |
ENST00000482108.1 ENST00000488574.1 |
PEG10 |
paternally expressed 10 |
| chr2_-_235405168 | 0.16 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr6_+_53659746 | 0.16 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr11_+_117947724 | 0.16 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr1_-_109203997 | 0.15 |
ENST00000370032.5 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr7_+_16793160 | 0.15 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr14_-_54421190 | 0.15 |
ENST00000417573.1 |
BMP4 |
bone morphogenetic protein 4 |
| chr11_+_45944190 | 0.15 |
ENST00000401752.1 ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B |
glycosyltransferase-like 1B |
| chr8_+_86089619 | 0.15 |
ENST00000256117.5 ENST00000416274.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr2_+_220491973 | 0.15 |
ENST00000358055.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr11_+_117947782 | 0.15 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr11_-_66139570 | 0.14 |
ENST00000311161.7 |
SLC29A2 |
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr15_+_90728145 | 0.14 |
ENST00000561085.1 ENST00000379122.3 ENST00000332496.6 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr2_+_95940220 | 0.14 |
ENST00000542147.1 |
PROM2 |
prominin 2 |
| chr10_-_101190202 | 0.14 |
ENST00000543866.1 ENST00000370508.5 |
GOT1 |
glutamic-oxaloacetic transaminase 1, soluble |
| chr22_-_18257178 | 0.14 |
ENST00000342111.5 |
BID |
BH3 interacting domain death agonist |
| chr8_-_124408652 | 0.14 |
ENST00000287394.5 |
ATAD2 |
ATPase family, AAA domain containing 2 |
| chr1_-_43751230 | 0.14 |
ENST00000523677.1 |
C1orf210 |
chromosome 1 open reading frame 210 |
| chr17_-_4463856 | 0.14 |
ENST00000574584.1 ENST00000381550.3 ENST00000301395.3 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr3_+_46283863 | 0.14 |
ENST00000545097.1 ENST00000541018.1 |
CCR3 |
chemokine (C-C motif) receptor 3 |
| chr8_+_31496809 | 0.14 |
ENST00000518104.1 ENST00000519301.1 |
NRG1 |
neuregulin 1 |
| chr3_-_128840604 | 0.13 |
ENST00000476465.1 ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43 |
RAB43, member RAS oncogene family |
| chr3_+_46283916 | 0.13 |
ENST00000395940.2 |
CCR3 |
chemokine (C-C motif) receptor 3 |
| chrX_+_102469997 | 0.13 |
ENST00000372695.5 ENST00000372691.3 |
BEX4 |
brain expressed, X-linked 4 |
| chr1_-_109203648 | 0.13 |
ENST00000370031.1 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr17_-_74533734 | 0.13 |
ENST00000589342.1 |
CYGB |
cytoglobin |
| chr17_+_73521763 | 0.12 |
ENST00000167462.5 ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2 |
lethal giant larvae homolog 2 (Drosophila) |
| chr11_+_1860832 | 0.12 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr11_+_1860682 | 0.12 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr2_-_163175133 | 0.12 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr19_-_14629224 | 0.12 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr16_+_202686 | 0.12 |
ENST00000252951.2 |
HBZ |
hemoglobin, zeta |
| chr17_-_4448361 | 0.12 |
ENST00000572759.1 |
MYBBP1A |
MYB binding protein (P160) 1a |
| chr3_-_49893958 | 0.12 |
ENST00000482243.1 ENST00000331456.2 ENST00000469027.1 |
TRAIP |
TRAF interacting protein |
| chr5_-_94620239 | 0.12 |
ENST00000515393.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr10_+_6779326 | 0.12 |
ENST00000417112.1 |
RP11-554I8.2 |
RP11-554I8.2 |
| chrX_+_48398053 | 0.12 |
ENST00000537536.1 ENST00000418627.1 |
TBC1D25 |
TBC1 domain family, member 25 |
| chr3_+_50649302 | 0.12 |
ENST00000446044.1 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
| chr14_+_56585048 | 0.12 |
ENST00000267460.4 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr8_+_86089460 | 0.11 |
ENST00000418930.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr15_-_83953466 | 0.11 |
ENST00000345382.2 |
BNC1 |
basonuclin 1 |
| chr1_+_150459873 | 0.11 |
ENST00000438568.2 ENST00000369054.2 ENST00000369064.3 ENST00000606933.1 |
TARS2 |
threonyl-tRNA synthetase 2, mitochondrial (putative) |
| chr6_+_41888926 | 0.11 |
ENST00000230340.4 |
BYSL |
bystin-like |
| chr19_+_33865218 | 0.11 |
ENST00000585933.2 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr1_-_160068465 | 0.11 |
ENST00000314485.7 ENST00000368086.1 |
IGSF8 |
immunoglobulin superfamily, member 8 |
| chr1_-_109203685 | 0.11 |
ENST00000402983.1 ENST00000420055.1 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr1_-_193029192 | 0.11 |
ENST00000417752.1 ENST00000367452.4 |
UCHL5 |
ubiquitin carboxyl-terminal hydrolase L5 |
| chr4_+_54966198 | 0.11 |
ENST00000326902.2 ENST00000503800.1 |
GSX2 |
GS homeobox 2 |
| chr3_-_121264848 | 0.10 |
ENST00000264233.5 |
POLQ |
polymerase (DNA directed), theta |
| chr16_-_23568651 | 0.10 |
ENST00000563232.1 ENST00000563459.1 ENST00000449606.1 |
EARS2 |
glutamyl-tRNA synthetase 2, mitochondrial |
| chr12_+_121647962 | 0.10 |
ENST00000542067.1 |
P2RX4 |
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr4_-_103997455 | 0.10 |
ENST00000362026.3 ENST00000503103.1 |
SLC9B2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr14_-_67878917 | 0.10 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr5_+_68788594 | 0.10 |
ENST00000396442.2 ENST00000380766.2 |
OCLN |
occludin |
| chr7_-_37024665 | 0.10 |
ENST00000396040.2 |
ELMO1 |
engulfment and cell motility 1 |
| chr2_-_31637560 | 0.10 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr6_-_163148700 | 0.10 |
ENST00000366894.1 ENST00000338468.3 |
PARK2 |
parkin RBR E3 ubiquitin protein ligase |
| chr17_+_4736627 | 0.10 |
ENST00000355280.6 ENST00000347992.7 |
MINK1 |
misshapen-like kinase 1 |
| chr2_+_230787201 | 0.09 |
ENST00000283946.3 |
FBXO36 |
F-box protein 36 |
| chr9_+_37485932 | 0.09 |
ENST00000377798.4 ENST00000442009.2 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr14_+_56584414 | 0.09 |
ENST00000559044.1 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr9_-_139137648 | 0.09 |
ENST00000358701.5 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
| chr10_+_104154229 | 0.09 |
ENST00000428099.1 ENST00000369966.3 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr2_-_54197915 | 0.09 |
ENST00000404125.1 |
PSME4 |
proteasome (prosome, macropain) activator subunit 4 |
| chr7_+_76109827 | 0.09 |
ENST00000446820.2 |
DTX2 |
deltex homolog 2 (Drosophila) |
| chr16_-_75498308 | 0.09 |
ENST00000569540.1 |
TMEM170A |
transmembrane protein 170A |
| chr6_-_73935163 | 0.09 |
ENST00000370388.3 |
KHDC1L |
KH homology domain containing 1-like |
| chr5_-_33892204 | 0.09 |
ENST00000504830.1 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr10_-_99393208 | 0.09 |
ENST00000307450.6 |
MORN4 |
MORN repeat containing 4 |
| chr5_+_149109825 | 0.09 |
ENST00000360453.4 ENST00000394320.3 ENST00000309241.5 |
PPARGC1B |
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr17_-_17109579 | 0.09 |
ENST00000321560.3 |
PLD6 |
phospholipase D family, member 6 |
| chrX_+_131157609 | 0.09 |
ENST00000496850.1 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr1_+_19578033 | 0.09 |
ENST00000330263.4 |
MRTO4 |
mRNA turnover 4 homolog (S. cerevisiae) |
| chr22_+_47070490 | 0.09 |
ENST00000408031.1 |
GRAMD4 |
GRAM domain containing 4 |
| chr18_+_43753974 | 0.09 |
ENST00000282059.6 ENST00000321319.6 |
C18orf25 |
chromosome 18 open reading frame 25 |
| chr17_-_42907564 | 0.09 |
ENST00000592524.1 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
| chr13_+_25254545 | 0.09 |
ENST00000218548.6 |
ATP12A |
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr8_+_99439214 | 0.09 |
ENST00000287042.4 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
| chr17_-_33775760 | 0.09 |
ENST00000534689.1 ENST00000532210.1 ENST00000526861.1 ENST00000531588.1 ENST00000285013.6 |
SLFN13 |
schlafen family member 13 |
| chr22_-_50913371 | 0.09 |
ENST00000348911.6 ENST00000380817.3 ENST00000390679.3 |
SBF1 |
SET binding factor 1 |
| chr16_-_69788816 | 0.09 |
ENST00000268802.5 |
NOB1 |
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr14_-_106622419 | 0.09 |
ENST00000390604.2 |
IGHV3-16 |
immunoglobulin heavy variable 3-16 (non-functional) |
| chr5_+_148724993 | 0.09 |
ENST00000513661.1 ENST00000329271.3 ENST00000416916.2 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
| chr6_-_35888824 | 0.09 |
ENST00000361690.3 ENST00000512445.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr19_+_51152702 | 0.09 |
ENST00000425202.1 |
C19orf81 |
chromosome 19 open reading frame 81 |
| chr7_+_136553370 | 0.09 |
ENST00000445907.2 |
CHRM2 |
cholinergic receptor, muscarinic 2 |
| chr9_+_37486005 | 0.09 |
ENST00000377792.3 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr2_+_230787213 | 0.09 |
ENST00000409992.1 |
FBXO36 |
F-box protein 36 |
| chr19_+_19496728 | 0.08 |
ENST00000537887.1 ENST00000417582.2 |
GATAD2A |
GATA zinc finger domain containing 2A |
| chr16_+_15068916 | 0.08 |
ENST00000455313.2 |
PDXDC1 |
pyridoxal-dependent decarboxylase domain containing 1 |
| chr1_-_205744205 | 0.08 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr3_+_196466710 | 0.08 |
ENST00000327134.3 |
PAK2 |
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr9_-_136006496 | 0.08 |
ENST00000372062.3 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
| chr1_-_151119087 | 0.08 |
ENST00000341697.3 ENST00000368914.3 |
SEMA6C |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr16_-_75498553 | 0.08 |
ENST00000569276.1 ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A RP11-77K12.1 |
transmembrane protein 170A Uncharacterized protein |
| chr17_-_39942940 | 0.08 |
ENST00000310706.5 ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP |
junction plakoglobin |
| chr9_+_138235095 | 0.08 |
ENST00000320778.2 |
C9orf62 |
chromosome 9 open reading frame 62 |
| chr10_+_5454505 | 0.08 |
ENST00000355029.4 |
NET1 |
neuroepithelial cell transforming 1 |
| chr3_+_38080691 | 0.08 |
ENST00000308059.6 ENST00000346219.3 ENST00000452631.2 |
DLEC1 |
deleted in lung and esophageal cancer 1 |
| chr18_-_4455260 | 0.08 |
ENST00000581527.1 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr5_+_61708582 | 0.08 |
ENST00000325324.6 |
IPO11 |
importin 11 |
| chr1_-_205744574 | 0.08 |
ENST00000367139.3 ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr10_-_99393242 | 0.08 |
ENST00000370635.3 ENST00000478953.1 ENST00000335628.3 |
MORN4 |
MORN repeat containing 4 |
| chr19_-_48867291 | 0.08 |
ENST00000435956.3 |
TMEM143 |
transmembrane protein 143 |
| chr15_+_40987327 | 0.08 |
ENST00000423169.2 ENST00000267868.3 ENST00000557850.1 ENST00000532743.1 ENST00000382643.3 |
RAD51 |
RAD51 recombinase |
| chr7_-_6010263 | 0.08 |
ENST00000455618.2 ENST00000405415.1 ENST00000404406.1 ENST00000542644.1 |
RSPH10B |
radial spoke head 10 homolog B (Chlamydomonas) |
| chr3_-_127842612 | 0.08 |
ENST00000417360.1 ENST00000322623.5 |
RUVBL1 |
RuvB-like AAA ATPase 1 |
| chr19_-_48867171 | 0.08 |
ENST00000377431.2 ENST00000436660.2 ENST00000541566.1 |
TMEM143 |
transmembrane protein 143 |
| chr5_+_56205878 | 0.08 |
ENST00000423328.1 |
SETD9 |
SET domain containing 9 |
| chr7_+_6793740 | 0.08 |
ENST00000403107.1 ENST00000404077.1 ENST00000435395.1 ENST00000418406.1 |
RSPH10B2 |
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chrX_+_131157290 | 0.08 |
ENST00000394334.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chrX_-_54070607 | 0.07 |
ENST00000338154.6 ENST00000338946.6 |
PHF8 |
PHD finger protein 8 |
| chr2_+_233897382 | 0.07 |
ENST00000233840.3 |
NEU2 |
sialidase 2 (cytosolic sialidase) |
| chr3_-_122102065 | 0.07 |
ENST00000479899.1 ENST00000291458.5 ENST00000497726.1 |
CCDC58 |
coiled-coil domain containing 58 |
| chr10_-_134145321 | 0.07 |
ENST00000368625.4 ENST00000368619.3 ENST00000456004.1 ENST00000368620.2 |
STK32C |
serine/threonine kinase 32C |
| chr14_-_51135036 | 0.07 |
ENST00000324679.4 |
SAV1 |
salvador homolog 1 (Drosophila) |
| chr3_+_45986511 | 0.07 |
ENST00000458629.1 ENST00000457814.1 |
CXCR6 |
chemokine (C-X-C motif) receptor 6 |
| chr3_+_113775576 | 0.07 |
ENST00000485050.1 ENST00000281273.4 |
QTRTD1 |
queuine tRNA-ribosyltransferase domain containing 1 |
| chr1_-_245027766 | 0.07 |
ENST00000283179.9 |
HNRNPU |
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr3_+_125687987 | 0.07 |
ENST00000514116.1 ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr16_-_66959429 | 0.07 |
ENST00000420652.1 ENST00000299759.6 |
RRAD |
Ras-related associated with diabetes |
| chr14_+_32546145 | 0.07 |
ENST00000556611.1 ENST00000539826.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr19_-_19384055 | 0.07 |
ENST00000389363.4 |
TM6SF2 |
transmembrane 6 superfamily member 2 |
| chr3_-_123710893 | 0.07 |
ENST00000467907.1 ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1 |
rhophilin associated tail protein 1 |
| chr14_-_53162361 | 0.07 |
ENST00000395686.3 |
ERO1L |
ERO1-like (S. cerevisiae) |
| chrX_+_131157322 | 0.07 |
ENST00000481105.1 ENST00000354719.6 ENST00000394335.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr14_+_32546274 | 0.07 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr16_+_31128978 | 0.07 |
ENST00000448516.2 ENST00000219797.4 |
KAT8 |
K(lysine) acetyltransferase 8 |
| chr1_+_3689325 | 0.07 |
ENST00000444870.2 ENST00000452264.1 |
SMIM1 |
small integral membrane protein 1 (Vel blood group) |
| chr1_-_33283754 | 0.07 |
ENST00000373477.4 |
YARS |
tyrosyl-tRNA synthetase |
| chr1_+_46806837 | 0.07 |
ENST00000537428.1 |
NSUN4 |
NOP2/Sun domain family, member 4 |
| chr22_+_31644388 | 0.07 |
ENST00000333611.4 ENST00000340552.4 |
LIMK2 |
LIM domain kinase 2 |
| chr6_+_79577189 | 0.07 |
ENST00000369940.2 |
IRAK1BP1 |
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr18_+_77439775 | 0.07 |
ENST00000299543.7 ENST00000075430.7 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr3_-_135914615 | 0.07 |
ENST00000309993.2 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
| chr6_+_168227611 | 0.07 |
ENST00000344191.4 ENST00000351017.4 ENST00000392108.3 ENST00000366806.2 ENST00000392112.1 ENST00000400824.4 ENST00000447894.2 ENST00000400822.3 |
MLLT4 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 |
| chr11_-_57103327 | 0.07 |
ENST00000529002.1 ENST00000278412.2 |
SSRP1 |
structure specific recognition protein 1 |
| chr8_-_8318847 | 0.07 |
ENST00000521218.1 |
CTA-398F10.2 |
CTA-398F10.2 |
| chr16_-_3086927 | 0.07 |
ENST00000572449.1 |
CCDC64B |
coiled-coil domain containing 64B |
| chr19_+_50084561 | 0.07 |
ENST00000246794.5 |
PRRG2 |
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr3_-_43147431 | 0.07 |
ENST00000441964.1 |
POMGNT2 |
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr1_-_1149506 | 0.07 |
ENST00000379236.3 |
TNFRSF4 |
tumor necrosis factor receptor superfamily, member 4 |
| chr1_-_93426998 | 0.07 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr21_-_47604318 | 0.07 |
ENST00000291672.5 ENST00000330205.6 |
SPATC1L |
spermatogenesis and centriole associated 1-like |
| chr1_-_78444738 | 0.06 |
ENST00000436586.2 ENST00000370768.2 |
FUBP1 |
far upstream element (FUSE) binding protein 1 |
| chrX_-_72299258 | 0.06 |
ENST00000453389.1 ENST00000373519.1 |
PABPC1L2A |
poly(A) binding protein, cytoplasmic 1-like 2A |
| chr11_+_93861993 | 0.06 |
ENST00000227638.3 ENST00000436171.2 |
PANX1 |
pannexin 1 |
| chr12_+_65563329 | 0.06 |
ENST00000308330.2 |
LEMD3 |
LEM domain containing 3 |
| chr18_-_4455283 | 0.06 |
ENST00000315677.3 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr6_-_163148780 | 0.06 |
ENST00000366892.1 ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2 |
parkin RBR E3 ubiquitin protein ligase |
| chr3_-_43147549 | 0.06 |
ENST00000344697.2 |
POMGNT2 |
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.5 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.3 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.2 | GO:0072097 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.3 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.1 | GO:0019858 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0060458 | right lung development(GO:0060458) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.3 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.0 | GO:1904253 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.0 | GO:0035568 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.0 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.0 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.0 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:0036446 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.0 | GO:0031982 | vesicle(GO:0031982) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.0 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0070700 | co-receptor binding(GO:0039706) BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.0 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |