Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for ESR1
Z-value: 1.02
Transcription factors associated with ESR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESR1
|
ENSG00000091831.17 | ESR1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESR1 | hg19_v2_chr6_+_152128371_152128454, hg19_v2_chr6_+_152011628_152011642, hg19_v2_chr6_+_152128686_152128803 | 0.15 | 7.3e-01 | Click! |
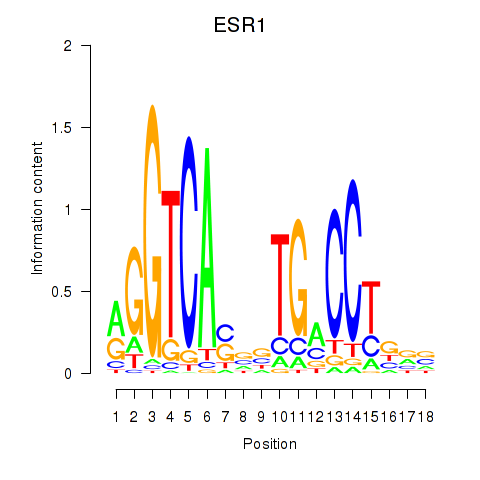
Activity profile of ESR1 motif
Sorted Z-values of ESR1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ESR1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39677971 | 0.70 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr18_+_21529811 | 0.70 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr13_-_20805109 | 0.63 |
ENST00000241124.6 |
GJB6 |
gap junction protein, beta 6, 30kDa |
| chr2_+_177015950 | 0.58 |
ENST00000306324.3 |
HOXD4 |
homeobox D4 |
| chr18_+_19749386 | 0.51 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr4_-_10023095 | 0.50 |
ENST00000264784.3 |
SLC2A9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr13_+_113548643 | 0.49 |
ENST00000375608.3 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr19_+_4279282 | 0.43 |
ENST00000599689.1 |
SHD |
Src homology 2 domain containing transforming protein D |
| chr5_-_139726181 | 0.43 |
ENST00000507104.1 ENST00000230990.6 |
HBEGF |
heparin-binding EGF-like growth factor |
| chr3_+_63897605 | 0.41 |
ENST00000487717.1 |
ATXN7 |
ataxin 7 |
| chr11_+_69924639 | 0.38 |
ENST00000538023.1 ENST00000398543.2 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr4_-_89080003 | 0.37 |
ENST00000237612.3 |
ABCG2 |
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr1_+_186798073 | 0.36 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr1_-_209825674 | 0.33 |
ENST00000367030.3 ENST00000356082.4 |
LAMB3 |
laminin, beta 3 |
| chr12_-_14721283 | 0.32 |
ENST00000240617.5 |
PLBD1 |
phospholipase B domain containing 1 |
| chr11_-_47270341 | 0.32 |
ENST00000529444.1 ENST00000530453.1 ENST00000537863.1 ENST00000529788.1 ENST00000444355.2 ENST00000527256.1 ENST00000529663.1 ENST00000256997.3 |
ACP2 |
acid phosphatase 2, lysosomal |
| chr21_-_46348626 | 0.31 |
ENST00000517563.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr5_-_16509101 | 0.30 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr9_+_130478345 | 0.30 |
ENST00000373289.3 ENST00000393748.4 |
TTC16 |
tetratricopeptide repeat domain 16 |
| chr16_+_66914264 | 0.30 |
ENST00000311765.2 ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr11_+_32851487 | 0.29 |
ENST00000257836.3 |
PRRG4 |
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr6_+_32121908 | 0.29 |
ENST00000375143.2 ENST00000424499.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr6_+_32121789 | 0.29 |
ENST00000437001.2 ENST00000375137.2 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr14_+_103566665 | 0.29 |
ENST00000559116.1 |
EXOC3L4 |
exocyst complex component 3-like 4 |
| chr12_-_42877726 | 0.29 |
ENST00000548696.1 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
| chr17_+_55173933 | 0.29 |
ENST00000539273.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr19_-_15090488 | 0.28 |
ENST00000594383.1 ENST00000598504.1 ENST00000597262.1 |
SLC1A6 |
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr12_-_95044309 | 0.27 |
ENST00000261226.4 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
| chr3_+_118865028 | 0.27 |
ENST00000460150.1 |
C3orf30 |
chromosome 3 open reading frame 30 |
| chr14_-_107078851 | 0.27 |
ENST00000390628.2 |
IGHV1-58 |
immunoglobulin heavy variable 1-58 |
| chr1_+_209848749 | 0.24 |
ENST00000367029.4 |
G0S2 |
G0/G1switch 2 |
| chr7_-_142139783 | 0.24 |
ENST00000390374.3 |
TRBV7-6 |
T cell receptor beta variable 7-6 |
| chr17_-_37323318 | 0.24 |
ENST00000444555.1 |
ARL5C |
ADP-ribosylation factor-like 5C |
| chr2_+_119981384 | 0.23 |
ENST00000393108.2 ENST00000354888.5 ENST00000450943.2 ENST00000393110.2 ENST00000393106.2 ENST00000409811.1 ENST00000393107.2 |
STEAP3 |
STEAP family member 3, metalloreductase |
| chr12_-_42877764 | 0.23 |
ENST00000455697.1 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
| chrX_+_38420623 | 0.23 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr19_-_54804173 | 0.23 |
ENST00000391744.3 ENST00000251390.3 |
LILRA3 |
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
| chr10_-_103880209 | 0.22 |
ENST00000425280.1 |
LDB1 |
LIM domain binding 1 |
| chr17_-_41277467 | 0.22 |
ENST00000494123.1 ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1 |
breast cancer 1, early onset |
| chr6_+_32121218 | 0.22 |
ENST00000414204.1 ENST00000361568.2 ENST00000395523.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr5_+_149109825 | 0.22 |
ENST00000360453.4 ENST00000394320.3 ENST00000309241.5 |
PPARGC1B |
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr13_+_37006398 | 0.21 |
ENST00000418263.1 |
CCNA1 |
cyclin A1 |
| chr19_+_58790314 | 0.21 |
ENST00000196548.5 ENST00000608843.1 |
ZNF8 ZNF8 |
Zinc finger protein 8 zinc finger protein 8 |
| chr21_-_47352477 | 0.21 |
ENST00000593412.1 |
PRED62 |
Uncharacterized protein |
| chr1_-_28503693 | 0.21 |
ENST00000373857.3 |
PTAFR |
platelet-activating factor receptor |
| chr3_+_148415571 | 0.21 |
ENST00000497524.1 ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr19_-_17375541 | 0.20 |
ENST00000252597.3 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr19_+_49838653 | 0.20 |
ENST00000598095.1 ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37 |
CD37 molecule |
| chr1_+_161677034 | 0.20 |
ENST00000349527.4 ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA |
Fc receptor-like A |
| chr11_-_1036706 | 0.20 |
ENST00000421673.2 |
MUC6 |
mucin 6, oligomeric mucus/gel-forming |
| chr1_+_153003671 | 0.20 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr16_-_10652993 | 0.20 |
ENST00000536829.1 |
EMP2 |
epithelial membrane protein 2 |
| chr22_+_46476192 | 0.20 |
ENST00000443490.1 |
FLJ27365 |
hsa-mir-4763 |
| chr16_+_56385290 | 0.20 |
ENST00000564727.1 |
GNAO1 |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr10_+_81466084 | 0.19 |
ENST00000342531.2 |
NUTM2B |
NUT family member 2B |
| chr12_+_125478241 | 0.19 |
ENST00000341446.8 |
BRI3BP |
BRI3 binding protein |
| chr2_+_177015122 | 0.19 |
ENST00000468418.3 |
HOXD3 |
homeobox D3 |
| chr1_-_246729544 | 0.19 |
ENST00000544618.1 ENST00000366514.4 |
TFB2M |
transcription factor B2, mitochondrial |
| chr1_-_223853348 | 0.19 |
ENST00000366872.5 |
CAPN8 |
calpain 8 |
| chr2_-_74779744 | 0.19 |
ENST00000409249.1 |
LOXL3 |
lysyl oxidase-like 3 |
| chr13_+_37005967 | 0.19 |
ENST00000440264.1 ENST00000449823.1 |
CCNA1 |
cyclin A1 |
| chr1_-_46089718 | 0.19 |
ENST00000421127.2 ENST00000343901.2 ENST00000528266.1 |
CCDC17 |
coiled-coil domain containing 17 |
| chr9_-_127263265 | 0.19 |
ENST00000373587.3 |
NR5A1 |
nuclear receptor subfamily 5, group A, member 1 |
| chr3_+_48488114 | 0.19 |
ENST00000421175.1 ENST00000320211.3 ENST00000346691.4 ENST00000357105.6 |
ATRIP |
ATR interacting protein |
| chr6_-_78173490 | 0.18 |
ENST00000369947.2 |
HTR1B |
5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
| chr20_+_43538756 | 0.18 |
ENST00000537323.1 ENST00000217073.2 |
PABPC1L |
poly(A) binding protein, cytoplasmic 1-like |
| chr1_-_153433120 | 0.18 |
ENST00000368723.3 |
S100A7 |
S100 calcium binding protein A7 |
| chr11_+_844406 | 0.18 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr13_+_37006421 | 0.18 |
ENST00000255465.4 |
CCNA1 |
cyclin A1 |
| chr15_+_90931450 | 0.18 |
ENST00000268182.5 ENST00000560738.1 ENST00000560418.1 |
IQGAP1 |
IQ motif containing GTPase activating protein 1 |
| chr8_+_75512010 | 0.18 |
ENST00000518190.1 ENST00000523118.1 |
RP11-758M4.1 |
Uncharacterized protein |
| chrX_-_72095808 | 0.18 |
ENST00000373529.5 |
DMRTC1 |
DMRT-like family C1 |
| chr14_-_106642049 | 0.18 |
ENST00000390605.2 |
IGHV1-18 |
immunoglobulin heavy variable 1-18 |
| chr2_-_98612350 | 0.18 |
ENST00000186436.5 |
TMEM131 |
transmembrane protein 131 |
| chr2_+_231090433 | 0.18 |
ENST00000486687.2 ENST00000350136.5 ENST00000392045.3 ENST00000417495.3 ENST00000343805.6 ENST00000420434.3 |
SP140 |
SP140 nuclear body protein |
| chr17_-_27507377 | 0.17 |
ENST00000531253.1 |
MYO18A |
myosin XVIIIA |
| chr7_-_122339162 | 0.17 |
ENST00000340112.2 |
RNF133 |
ring finger protein 133 |
| chr11_-_118134997 | 0.17 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr16_-_74734672 | 0.17 |
ENST00000306247.7 ENST00000575686.1 |
MLKL |
mixed lineage kinase domain-like |
| chr17_-_27507395 | 0.17 |
ENST00000354329.4 ENST00000527372.1 |
MYO18A |
myosin XVIIIA |
| chr4_-_3534139 | 0.17 |
ENST00000500728.2 |
LRPAP1 |
low density lipoprotein receptor-related protein associated protein 1 |
| chr11_+_33563821 | 0.17 |
ENST00000321505.4 ENST00000265654.5 ENST00000389726.3 |
KIAA1549L |
KIAA1549-like |
| chr19_-_49176264 | 0.17 |
ENST00000270235.4 ENST00000596844.1 |
NTN5 |
netrin 5 |
| chr4_-_101111615 | 0.17 |
ENST00000273990.2 |
DDIT4L |
DNA-damage-inducible transcript 4-like |
| chr20_-_4982132 | 0.16 |
ENST00000338244.1 ENST00000424750.2 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr6_-_136871957 | 0.16 |
ENST00000354570.3 |
MAP7 |
microtubule-associated protein 7 |
| chr11_+_94277017 | 0.16 |
ENST00000358752.2 |
FUT4 |
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr22_+_32455111 | 0.16 |
ENST00000543737.1 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr4_-_103266219 | 0.16 |
ENST00000394833.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr15_-_49103235 | 0.16 |
ENST00000380950.2 |
CEP152 |
centrosomal protein 152kDa |
| chr15_+_81426588 | 0.16 |
ENST00000286732.4 |
C15orf26 |
chromosome 15 open reading frame 26 |
| chr1_+_31885963 | 0.16 |
ENST00000373709.3 |
SERINC2 |
serine incorporator 2 |
| chr17_+_30677136 | 0.16 |
ENST00000394670.4 ENST00000321233.6 ENST00000394673.2 ENST00000341711.6 ENST00000579634.1 ENST00000580759.1 ENST00000342555.6 ENST00000577908.1 ENST00000394679.5 ENST00000582165.1 |
ZNF207 |
zinc finger protein 207 |
| chr6_-_105584560 | 0.15 |
ENST00000336775.5 |
BVES |
blood vessel epicardial substance |
| chr9_+_139847347 | 0.15 |
ENST00000371632.3 |
LCN12 |
lipocalin 12 |
| chr8_-_144691718 | 0.15 |
ENST00000377579.3 ENST00000433751.1 ENST00000220966.6 |
PYCRL |
pyrroline-5-carboxylate reductase-like |
| chr17_+_7323634 | 0.15 |
ENST00000323675.3 |
SPEM1 |
spermatid maturation 1 |
| chr22_+_22676808 | 0.15 |
ENST00000390290.2 |
IGLV1-51 |
immunoglobulin lambda variable 1-51 |
| chr1_+_34632484 | 0.15 |
ENST00000373374.3 |
C1orf94 |
chromosome 1 open reading frame 94 |
| chr10_+_54074033 | 0.15 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr19_+_46002868 | 0.15 |
ENST00000396735.2 |
PPM1N |
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr14_+_75536280 | 0.15 |
ENST00000238686.8 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chr12_-_51420128 | 0.15 |
ENST00000262051.7 ENST00000547732.1 ENST00000262052.5 ENST00000546488.1 ENST00000550714.1 ENST00000548193.1 ENST00000547579.1 ENST00000546743.1 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr11_+_117947724 | 0.15 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr17_+_57784826 | 0.14 |
ENST00000262291.4 |
VMP1 |
vacuole membrane protein 1 |
| chr16_-_74734742 | 0.14 |
ENST00000308807.7 ENST00000573267.1 |
MLKL |
mixed lineage kinase domain-like |
| chr15_+_33010175 | 0.14 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr9_-_34637806 | 0.14 |
ENST00000477726.1 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr19_-_51512804 | 0.14 |
ENST00000594211.1 ENST00000376832.4 |
KLK9 |
kallikrein-related peptidase 9 |
| chr21_-_46293644 | 0.14 |
ENST00000330938.3 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
| chr7_-_134001663 | 0.14 |
ENST00000378509.4 |
SLC35B4 |
solute carrier family 35 (UDP-xylose/UDP-N-acetylglucosamine transporter), member B4 |
| chr1_+_901847 | 0.14 |
ENST00000379410.3 ENST00000379409.2 ENST00000379407.3 |
PLEKHN1 |
pleckstrin homology domain containing, family N member 1 |
| chr1_-_205819245 | 0.14 |
ENST00000367136.4 |
PM20D1 |
peptidase M20 domain containing 1 |
| chr16_+_29690358 | 0.14 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr9_-_34637718 | 0.14 |
ENST00000378892.1 ENST00000277010.4 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr1_+_23037323 | 0.14 |
ENST00000544305.1 ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2 |
EPH receptor B2 |
| chr14_+_75536335 | 0.14 |
ENST00000554763.1 ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chr21_-_46293586 | 0.14 |
ENST00000445724.2 ENST00000397887.3 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
| chr6_+_144164455 | 0.14 |
ENST00000367576.5 |
LTV1 |
LTV1 homolog (S. cerevisiae) |
| chr17_+_37793318 | 0.14 |
ENST00000336308.5 |
STARD3 |
StAR-related lipid transfer (START) domain containing 3 |
| chr14_+_23340822 | 0.14 |
ENST00000359591.4 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
| chr9_-_139927462 | 0.14 |
ENST00000314412.6 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
| chr21_-_40817645 | 0.14 |
ENST00000438404.1 ENST00000358268.2 ENST00000411566.1 ENST00000451131.1 ENST00000418018.1 ENST00000415863.1 ENST00000426783.1 ENST00000288350.3 ENST00000485895.2 ENST00000448288.2 ENST00000456017.1 ENST00000434281.1 |
LCA5L |
Leber congenital amaurosis 5-like |
| chr20_+_44098385 | 0.14 |
ENST00000217425.5 ENST00000339946.3 |
WFDC2 |
WAP four-disulfide core domain 2 |
| chr15_+_51973680 | 0.14 |
ENST00000542355.2 |
SCG3 |
secretogranin III |
| chr11_-_118135160 | 0.13 |
ENST00000438295.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr8_+_120885949 | 0.13 |
ENST00000523492.1 ENST00000286234.5 |
DEPTOR |
DEP domain containing MTOR-interacting protein |
| chr12_+_123259063 | 0.13 |
ENST00000392441.4 ENST00000539171.1 |
CCDC62 |
coiled-coil domain containing 62 |
| chr7_-_150884873 | 0.13 |
ENST00000377867.3 |
ASB10 |
ankyrin repeat and SOCS box containing 10 |
| chr3_-_93781750 | 0.13 |
ENST00000314636.2 |
DHFRL1 |
dihydrofolate reductase-like 1 |
| chr19_-_54761148 | 0.13 |
ENST00000316219.5 |
LILRB5 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 5 |
| chr8_-_144099795 | 0.13 |
ENST00000522060.1 ENST00000517833.1 ENST00000502167.2 ENST00000518831.1 |
RP11-273G15.2 |
RP11-273G15.2 |
| chr11_+_64008525 | 0.13 |
ENST00000449942.2 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr19_+_45349630 | 0.13 |
ENST00000252483.5 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr6_-_105585022 | 0.13 |
ENST00000314641.5 |
BVES |
blood vessel epicardial substance |
| chr22_+_50312316 | 0.13 |
ENST00000328268.4 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chr19_-_22806764 | 0.13 |
ENST00000598042.1 |
AC011516.2 |
AC011516.2 |
| chr4_+_15376165 | 0.13 |
ENST00000382383.3 ENST00000429690.1 |
C1QTNF7 |
C1q and tumor necrosis factor related protein 7 |
| chr10_-_27444143 | 0.13 |
ENST00000477432.1 |
YME1L1 |
YME1-like 1 ATPase |
| chr6_-_44225231 | 0.12 |
ENST00000538577.1 ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr11_+_827553 | 0.12 |
ENST00000528542.2 ENST00000450448.1 |
EFCAB4A |
EF-hand calcium binding domain 4A |
| chr19_+_50194360 | 0.12 |
ENST00000323446.5 ENST00000392518.4 ENST00000598396.1 ENST00000598293.1 ENST00000354199.5 ENST00000405931.2 ENST00000602019.1 |
CPT1C |
carnitine palmitoyltransferase 1C |
| chr19_-_6720686 | 0.12 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr7_-_98030360 | 0.12 |
ENST00000005260.8 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
| chr19_+_42300548 | 0.12 |
ENST00000344550.4 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr3_-_33759699 | 0.12 |
ENST00000399362.4 ENST00000359576.5 ENST00000307312.7 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr22_+_23154239 | 0.12 |
ENST00000390315.2 |
IGLV3-10 |
immunoglobulin lambda variable 3-10 |
| chr10_-_27389392 | 0.12 |
ENST00000376087.4 |
ANKRD26 |
ankyrin repeat domain 26 |
| chr19_+_44116236 | 0.12 |
ENST00000417606.1 |
SRRM5 |
serine/arginine repetitive matrix 5 |
| chr12_-_51420108 | 0.12 |
ENST00000547198.1 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr15_-_75017711 | 0.12 |
ENST00000567032.1 ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1 |
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr19_-_17375527 | 0.12 |
ENST00000431146.2 ENST00000594190.1 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr17_+_65040678 | 0.12 |
ENST00000226021.3 |
CACNG1 |
calcium channel, voltage-dependent, gamma subunit 1 |
| chr2_+_128848881 | 0.12 |
ENST00000259253.6 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr2_+_128848740 | 0.12 |
ENST00000375990.3 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr17_-_57784755 | 0.12 |
ENST00000537860.1 ENST00000393038.2 ENST00000409433.2 |
PTRH2 |
peptidyl-tRNA hydrolase 2 |
| chr1_-_155990580 | 0.12 |
ENST00000531917.1 ENST00000480567.1 ENST00000526212.1 ENST00000529008.1 ENST00000496742.1 ENST00000295702.4 |
SSR2 |
signal sequence receptor, beta (translocon-associated protein beta) |
| chr19_-_54761080 | 0.12 |
ENST00000345866.6 ENST00000449561.2 ENST00000450632.1 |
LILRB5 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 5 |
| chr9_-_115653176 | 0.12 |
ENST00000374228.4 |
SLC46A2 |
solute carrier family 46, member 2 |
| chr2_+_113479063 | 0.12 |
ENST00000327581.4 |
NT5DC4 |
5'-nucleotidase domain containing 4 |
| chr22_+_22681656 | 0.12 |
ENST00000390291.2 |
IGLV1-50 |
immunoglobulin lambda variable 1-50 (non-functional) |
| chr12_-_48119301 | 0.12 |
ENST00000545824.2 ENST00000422538.3 |
ENDOU |
endonuclease, polyU-specific |
| chr8_-_67874805 | 0.12 |
ENST00000563496.1 |
TCF24 |
transcription factor 24 |
| chr1_+_1167594 | 0.12 |
ENST00000379198.2 |
B3GALT6 |
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr15_+_50474412 | 0.12 |
ENST00000380902.4 |
SLC27A2 |
solute carrier family 27 (fatty acid transporter), member 2 |
| chr11_+_17756279 | 0.12 |
ENST00000265969.6 |
KCNC1 |
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr3_-_125655882 | 0.12 |
ENST00000340333.3 |
ALG1L |
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase-like |
| chr15_+_50474385 | 0.12 |
ENST00000267842.5 |
SLC27A2 |
solute carrier family 27 (fatty acid transporter), member 2 |
| chr17_+_57784997 | 0.12 |
ENST00000537567.1 ENST00000539763.1 ENST00000587945.1 ENST00000536180.1 ENST00000589823.2 ENST00000592106.1 ENST00000591315.1 ENST00000545362.1 |
VMP1 |
vacuole membrane protein 1 |
| chr3_-_33759541 | 0.12 |
ENST00000468888.2 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr17_+_26684604 | 0.12 |
ENST00000292114.3 ENST00000509083.1 |
TMEM199 |
transmembrane protein 199 |
| chr11_+_64008443 | 0.12 |
ENST00000309366.4 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr6_+_31462658 | 0.12 |
ENST00000538442.1 |
MICB |
MHC class I polypeptide-related sequence B |
| chr12_-_48119340 | 0.12 |
ENST00000229003.3 |
ENDOU |
endonuclease, polyU-specific |
| chr11_+_117947782 | 0.12 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr11_+_844067 | 0.12 |
ENST00000397406.1 ENST00000409543.2 ENST00000525201.1 |
TSPAN4 |
tetraspanin 4 |
| chr1_+_207070775 | 0.12 |
ENST00000391929.3 ENST00000294984.2 ENST00000367093.3 |
IL24 |
interleukin 24 |
| chr13_-_103451307 | 0.12 |
ENST00000376004.4 |
KDELC1 |
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr11_+_66624527 | 0.11 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr8_+_142138711 | 0.11 |
ENST00000518347.1 ENST00000262585.2 ENST00000424248.1 ENST00000519811.1 ENST00000520986.1 ENST00000523058.1 |
DENND3 |
DENN/MADD domain containing 3 |
| chr10_+_101419187 | 0.11 |
ENST00000370489.4 |
ENTPD7 |
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr16_-_28857677 | 0.11 |
ENST00000313511.3 |
TUFM |
Tu translation elongation factor, mitochondrial |
| chr17_+_37793378 | 0.11 |
ENST00000544210.2 ENST00000581894.1 ENST00000394250.4 ENST00000579479.1 ENST00000577248.1 ENST00000580611.1 |
STARD3 |
StAR-related lipid transfer (START) domain containing 3 |
| chr2_+_204103663 | 0.11 |
ENST00000356079.4 ENST00000429815.2 |
CYP20A1 |
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr11_+_43380459 | 0.11 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chr19_+_55315087 | 0.11 |
ENST00000345540.5 ENST00000357494.4 ENST00000396293.1 ENST00000346587.4 ENST00000396289.5 |
KIR2DL4 |
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4 |
| chr1_+_207925391 | 0.11 |
ENST00000358170.2 ENST00000354848.1 ENST00000322918.5 ENST00000367042.1 ENST00000367041.1 ENST00000357714.1 ENST00000322875.4 ENST00000367047.1 ENST00000441839.2 ENST00000361067.1 ENST00000360212.2 ENST00000480003.1 |
CD46 |
CD46 molecule, complement regulatory protein |
| chr1_+_1217489 | 0.11 |
ENST00000325425.8 ENST00000400928.3 |
SCNN1D |
sodium channel, non-voltage-gated 1, delta subunit |
| chr12_-_51419924 | 0.11 |
ENST00000541174.2 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr1_+_179851823 | 0.11 |
ENST00000435319.4 |
TOR1AIP1 |
torsin A interacting protein 1 |
| chr3_+_58223228 | 0.11 |
ENST00000478253.1 ENST00000295962.4 |
ABHD6 |
abhydrolase domain containing 6 |
| chr2_-_233415220 | 0.11 |
ENST00000408957.3 |
TIGD1 |
tigger transposable element derived 1 |
| chr1_-_16763685 | 0.11 |
ENST00000540400.1 |
SPATA21 |
spermatogenesis associated 21 |
| chr10_-_103578162 | 0.11 |
ENST00000361464.3 ENST00000357797.5 ENST00000370094.3 |
MGEA5 |
meningioma expressed antigen 5 (hyaluronidase) |
| chr17_-_3337135 | 0.11 |
ENST00000248384.1 |
OR1E2 |
olfactory receptor, family 1, subfamily E, member 2 |
| chr20_+_58296265 | 0.11 |
ENST00000395636.2 ENST00000361300.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr12_-_88535747 | 0.11 |
ENST00000309041.7 |
CEP290 |
centrosomal protein 290kDa |
| chr1_+_236958554 | 0.11 |
ENST00000366577.5 ENST00000418145.2 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr19_+_45349432 | 0.10 |
ENST00000252485.4 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr19_-_42498231 | 0.10 |
ENST00000602133.1 |
ATP1A3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.5 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.3 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.2 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 0.8 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.2 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.1 | 0.2 | GO:1902565 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.2 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.1 | GO:1900158 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.3 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.3 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.2 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.0 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:1900738 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.0 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.1 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.1 | GO:0060374 | positive regulation of neutrophil apoptotic process(GO:0033031) mast cell differentiation(GO:0060374) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.0 | GO:1901671 | L-ascorbic acid biosynthetic process(GO:0019853) positive regulation of superoxide dismutase activity(GO:1901671) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.0 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) |
| 0.0 | 0.6 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0099638 | endosome to plasma membrane protein transport(GO:0099638) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0098712 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) L-glutamate import across plasma membrane(GO:0098712) |
| 0.0 | 0.0 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.2 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0071745 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.0 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.1 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0008523 | L-ascorbate:sodium symporter activity(GO:0008520) sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.2 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.0 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.0 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |