Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for ETV3
Z-value: 0.59
Transcription factors associated with ETV3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV3
|
ENSG00000117036.7 | ETV3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV3 | hg19_v2_chr1_-_157108130_157108173 | 0.53 | 1.8e-01 | Click! |
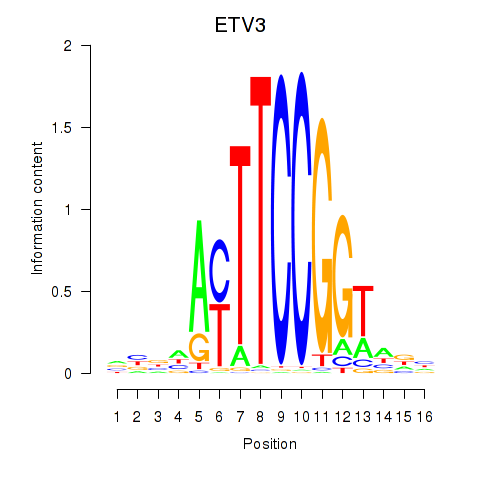
Activity profile of ETV3 motif
Sorted Z-values of ETV3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51522955 | 0.89 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr11_-_118122996 | 0.74 |
ENST00000525386.1 ENST00000527472.1 ENST00000278949.4 |
MPZL3 |
myelin protein zero-like 3 |
| chr19_-_51523412 | 0.57 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr19_-_51523275 | 0.53 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr4_+_40198527 | 0.53 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr2_+_118846008 | 0.43 |
ENST00000245787.4 |
INSIG2 |
insulin induced gene 2 |
| chr6_+_28048753 | 0.41 |
ENST00000377325.1 |
ZNF165 |
zinc finger protein 165 |
| chr11_+_61129456 | 0.39 |
ENST00000278826.6 |
TMEM138 |
transmembrane protein 138 |
| chr2_+_32502952 | 0.38 |
ENST00000238831.4 |
YIPF4 |
Yip1 domain family, member 4 |
| chr21_+_18885430 | 0.34 |
ENST00000356275.6 ENST00000400165.1 ENST00000400169.1 ENST00000306618.10 |
CXADR |
coxsackie virus and adenovirus receptor |
| chr21_+_18885318 | 0.32 |
ENST00000400166.1 |
CXADR |
coxsackie virus and adenovirus receptor |
| chr1_-_161015752 | 0.30 |
ENST00000435396.1 ENST00000368021.3 |
USF1 |
upstream transcription factor 1 |
| chr11_-_61129335 | 0.29 |
ENST00000545361.1 ENST00000539128.1 ENST00000546151.1 ENST00000447532.2 |
CYB561A3 |
cytochrome b561 family, member A3 |
| chr11_-_61129723 | 0.25 |
ENST00000537680.1 ENST00000426130.2 ENST00000294072.4 |
CYB561A3 |
cytochrome b561 family, member A3 |
| chr6_-_28554977 | 0.25 |
ENST00000452236.2 |
SCAND3 |
SCAN domain containing 3 |
| chr15_+_74833518 | 0.25 |
ENST00000346246.5 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
| chr11_-_9336234 | 0.24 |
ENST00000528080.1 |
TMEM41B |
transmembrane protein 41B |
| chr11_+_63606477 | 0.24 |
ENST00000508192.1 ENST00000361128.5 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr11_+_63606373 | 0.23 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr19_-_19774473 | 0.23 |
ENST00000357324.6 |
ATP13A1 |
ATPase type 13A1 |
| chr15_+_77287426 | 0.22 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr9_-_33473882 | 0.21 |
ENST00000455041.2 ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6 |
nucleolar protein 6 (RNA-associated) |
| chr9_-_86571628 | 0.21 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr16_-_4466622 | 0.21 |
ENST00000570645.1 ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7 |
coronin 7 |
| chr11_+_63606558 | 0.20 |
ENST00000350490.7 ENST00000502399.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr12_+_9102632 | 0.20 |
ENST00000539240.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr16_-_23652570 | 0.20 |
ENST00000261584.4 |
PALB2 |
partner and localizer of BRCA2 |
| chr22_-_38245304 | 0.20 |
ENST00000609454.1 |
ANKRD54 |
ankyrin repeat domain 54 |
| chr1_-_20987851 | 0.20 |
ENST00000464364.1 ENST00000602624.2 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr17_+_7387677 | 0.19 |
ENST00000322644.6 |
POLR2A |
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr8_-_56685966 | 0.19 |
ENST00000334667.2 |
TMEM68 |
transmembrane protein 68 |
| chr1_-_20987982 | 0.19 |
ENST00000375048.3 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr3_+_57541975 | 0.19 |
ENST00000487257.1 ENST00000311180.8 |
PDE12 |
phosphodiesterase 12 |
| chr12_+_65563329 | 0.19 |
ENST00000308330.2 |
LEMD3 |
LEM domain containing 3 |
| chr8_-_56685859 | 0.19 |
ENST00000523423.1 ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68 |
transmembrane protein 68 |
| chr2_+_9696028 | 0.19 |
ENST00000607241.1 |
RP11-214N9.1 |
RP11-214N9.1 |
| chr11_+_118230287 | 0.19 |
ENST00000252108.3 ENST00000431736.2 |
UBE4A |
ubiquitination factor E4A |
| chr6_-_33239612 | 0.18 |
ENST00000482399.1 ENST00000445902.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr6_-_86303523 | 0.18 |
ENST00000513865.1 ENST00000369627.2 ENST00000514419.1 ENST00000509338.1 ENST00000314673.3 ENST00000346348.3 |
SNX14 |
sorting nexin 14 |
| chr1_-_38273840 | 0.18 |
ENST00000373044.2 |
YRDC |
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr18_+_72265084 | 0.18 |
ENST00000582337.1 |
ZNF407 |
zinc finger protein 407 |
| chr17_-_39093672 | 0.18 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chr1_-_20987889 | 0.18 |
ENST00000415136.2 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr2_-_9695847 | 0.18 |
ENST00000310823.3 ENST00000497134.1 |
ADAM17 |
ADAM metallopeptidase domain 17 |
| chr8_+_146052849 | 0.18 |
ENST00000532777.1 ENST00000325241.6 ENST00000446747.2 ENST00000525266.1 ENST00000544249.1 ENST00000325217.5 ENST00000533314.1 ENST00000527218.1 ENST00000529819.1 ENST00000528372.1 |
ZNF7 |
zinc finger protein 7 |
| chr11_+_100558384 | 0.18 |
ENST00000524892.2 ENST00000298815.8 |
ARHGAP42 |
Rho GTPase activating protein 42 |
| chr15_+_43663257 | 0.17 |
ENST00000260383.7 ENST00000564079.1 |
TUBGCP4 |
tubulin, gamma complex associated protein 4 |
| chr19_-_16653226 | 0.17 |
ENST00000198939.6 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr13_-_46626847 | 0.17 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr17_-_57784755 | 0.17 |
ENST00000537860.1 ENST00000393038.2 ENST00000409433.2 |
PTRH2 |
peptidyl-tRNA hydrolase 2 |
| chr16_+_67261008 | 0.17 |
ENST00000304800.9 ENST00000563953.1 ENST00000565201.1 |
TMEM208 |
transmembrane protein 208 |
| chr2_+_148602058 | 0.17 |
ENST00000241416.7 ENST00000535787.1 ENST00000404590.1 |
ACVR2A |
activin A receptor, type IIA |
| chr5_+_149737202 | 0.17 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
| chr11_-_64885111 | 0.17 |
ENST00000528598.1 ENST00000310597.4 |
ZNHIT2 |
zinc finger, HIT-type containing 2 |
| chr6_-_166755995 | 0.17 |
ENST00000361731.3 |
SFT2D1 |
SFT2 domain containing 1 |
| chr14_-_75643296 | 0.17 |
ENST00000303575.4 |
TMED10 |
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr1_-_92952433 | 0.16 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr19_-_16653325 | 0.16 |
ENST00000546361.2 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr2_-_75788038 | 0.16 |
ENST00000393913.3 ENST00000410113.1 |
EVA1A |
eva-1 homolog A (C. elegans) |
| chr17_+_7211280 | 0.16 |
ENST00000419711.2 ENST00000571955.1 ENST00000573714.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr1_+_100315613 | 0.16 |
ENST00000361915.3 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr22_+_38004473 | 0.16 |
ENST00000414350.3 ENST00000343632.4 |
GGA1 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr21_+_45527171 | 0.16 |
ENST00000291576.7 ENST00000456705.1 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
| chr1_-_156698591 | 0.16 |
ENST00000368219.1 |
ISG20L2 |
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr19_-_19626838 | 0.15 |
ENST00000360913.3 |
TSSK6 |
testis-specific serine kinase 6 |
| chr12_-_123215306 | 0.15 |
ENST00000356987.2 ENST00000436083.2 |
HCAR1 |
hydroxycarboxylic acid receptor 1 |
| chr15_+_43425672 | 0.15 |
ENST00000260403.2 |
TMEM62 |
transmembrane protein 62 |
| chr6_+_144164455 | 0.15 |
ENST00000367576.5 |
LTV1 |
LTV1 homolog (S. cerevisiae) |
| chr1_-_156698181 | 0.15 |
ENST00000313146.6 |
ISG20L2 |
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr8_+_145137489 | 0.15 |
ENST00000355091.4 ENST00000525087.1 ENST00000361036.6 ENST00000524418.1 |
GPAA1 |
glycosylphosphatidylinositol anchor attachment 1 |
| chr1_-_160001737 | 0.15 |
ENST00000368090.2 |
PIGM |
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr2_+_99225018 | 0.15 |
ENST00000357765.2 ENST00000409975.1 |
UNC50 |
unc-50 homolog (C. elegans) |
| chr10_+_105156364 | 0.15 |
ENST00000369797.3 |
PDCD11 |
programmed cell death 11 |
| chr6_-_31620403 | 0.15 |
ENST00000451898.1 ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chrX_+_54556633 | 0.14 |
ENST00000336470.4 ENST00000360845.2 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr8_+_95565947 | 0.14 |
ENST00000523011.1 |
RP11-267M23.4 |
RP11-267M23.4 |
| chr17_-_29641104 | 0.14 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr5_+_79703823 | 0.14 |
ENST00000338008.5 ENST00000510158.1 ENST00000505560.1 |
ZFYVE16 |
zinc finger, FYVE domain containing 16 |
| chr6_+_43484760 | 0.14 |
ENST00000372389.3 ENST00000372344.2 ENST00000304004.3 ENST00000423780.1 |
POLR1C |
polymerase (RNA) I polypeptide C, 30kDa |
| chr1_+_44435646 | 0.14 |
ENST00000255108.3 ENST00000412950.2 ENST00000396758.2 |
DPH2 |
DPH2 homolog (S. cerevisiae) |
| chr10_-_96122682 | 0.14 |
ENST00000371361.3 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr18_+_21032781 | 0.13 |
ENST00000339486.3 |
RIOK3 |
RIO kinase 3 |
| chr1_+_169337172 | 0.13 |
ENST00000367807.3 ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1 |
basic leucine zipper nuclear factor 1 |
| chr14_-_68066975 | 0.13 |
ENST00000559581.1 ENST00000560722.1 ENST00000559415.1 ENST00000216452.4 |
PIGH |
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr12_+_132195617 | 0.13 |
ENST00000261674.4 ENST00000535236.1 ENST00000541286.1 |
SFSWAP |
splicing factor, suppressor of white-apricot homolog (Drosophila) |
| chr12_+_51632666 | 0.13 |
ENST00000604900.1 |
DAZAP2 |
DAZ associated protein 2 |
| chr1_-_32687923 | 0.13 |
ENST00000309777.6 ENST00000344461.3 ENST00000373593.1 ENST00000545122.1 |
TMEM234 |
transmembrane protein 234 |
| chr21_-_36260980 | 0.13 |
ENST00000344691.4 ENST00000358356.5 |
RUNX1 |
runt-related transcription factor 1 |
| chr22_+_46731596 | 0.13 |
ENST00000381019.3 |
TRMU |
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr16_+_14726672 | 0.13 |
ENST00000261658.2 ENST00000563971.1 |
BFAR |
bifunctional apoptosis regulator |
| chr14_-_68066849 | 0.13 |
ENST00000558493.1 ENST00000561272.1 |
PIGH |
phosphatidylinositol glycan anchor biosynthesis, class H |
| chrX_-_27417088 | 0.13 |
ENST00000608735.1 ENST00000422048.1 |
RP11-268G12.1 |
RP11-268G12.1 |
| chr5_+_99871004 | 0.12 |
ENST00000312637.4 |
FAM174A |
family with sequence similarity 174, member A |
| chr19_-_55791431 | 0.12 |
ENST00000593263.1 ENST00000376343.3 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr6_-_33239712 | 0.12 |
ENST00000436044.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr11_+_61891445 | 0.12 |
ENST00000394818.3 ENST00000533896.1 ENST00000278849.4 |
INCENP |
inner centromere protein antigens 135/155kDa |
| chr20_-_44993012 | 0.12 |
ENST00000372229.1 ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2 |
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr11_-_62389449 | 0.12 |
ENST00000534026.1 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr15_-_43398274 | 0.12 |
ENST00000382177.2 ENST00000290650.4 |
UBR1 |
ubiquitin protein ligase E3 component n-recognin 1 |
| chr19_-_11039188 | 0.12 |
ENST00000588347.1 |
YIPF2 |
Yip1 domain family, member 2 |
| chr17_+_57784826 | 0.12 |
ENST00000262291.4 |
VMP1 |
vacuole membrane protein 1 |
| chr13_+_100153665 | 0.12 |
ENST00000376387.4 |
TM9SF2 |
transmembrane 9 superfamily member 2 |
| chr11_+_46722368 | 0.12 |
ENST00000311764.2 |
ZNF408 |
zinc finger protein 408 |
| chr2_+_177134134 | 0.11 |
ENST00000249442.6 ENST00000392529.2 ENST00000443241.1 |
MTX2 |
metaxin 2 |
| chr19_-_55690758 | 0.11 |
ENST00000590851.1 |
SYT5 |
synaptotagmin V |
| chr12_-_49110613 | 0.11 |
ENST00000261900.3 |
CCNT1 |
cyclin T1 |
| chr22_+_38004832 | 0.11 |
ENST00000405147.3 ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr17_-_7155274 | 0.11 |
ENST00000318988.6 ENST00000575783.1 ENST00000573600.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr17_+_7211656 | 0.11 |
ENST00000416016.2 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr2_+_177134201 | 0.11 |
ENST00000452865.1 |
MTX2 |
metaxin 2 |
| chr19_+_11546440 | 0.11 |
ENST00000589126.1 ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr13_-_96705624 | 0.11 |
ENST00000376747.3 ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2 |
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr14_+_69865401 | 0.11 |
ENST00000556605.1 ENST00000336643.5 ENST00000031146.4 |
SLC39A9 |
solute carrier family 39, member 9 |
| chr19_+_797443 | 0.11 |
ENST00000394601.4 ENST00000589575.1 |
PTBP1 |
polypyrimidine tract binding protein 1 |
| chr3_+_142720366 | 0.11 |
ENST00000493782.1 ENST00000397933.2 ENST00000473835.2 ENST00000493598.2 |
U2SURP |
U2 snRNP-associated SURP domain containing |
| chr11_-_71791435 | 0.11 |
ENST00000351960.6 ENST00000541719.1 ENST00000535111.1 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
| chr11_+_65819802 | 0.11 |
ENST00000528302.1 ENST00000322535.6 ENST00000524627.1 ENST00000533595.1 ENST00000530322.1 |
SF3B2 |
splicing factor 3b, subunit 2, 145kDa |
| chr18_-_47813940 | 0.11 |
ENST00000586837.1 ENST00000412036.2 ENST00000589940.1 |
CXXC1 |
CXXC finger protein 1 |
| chr10_-_14996017 | 0.11 |
ENST00000378241.1 ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C |
DNA cross-link repair 1C |
| chr12_+_21590549 | 0.11 |
ENST00000545178.1 ENST00000240651.9 |
PYROXD1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr11_-_134093827 | 0.11 |
ENST00000534548.2 |
NCAPD3 |
non-SMC condensin II complex, subunit D3 |
| chr16_+_70557685 | 0.11 |
ENST00000302516.5 ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3 |
splicing factor 3b, subunit 3, 130kDa |
| chr17_+_57784997 | 0.11 |
ENST00000537567.1 ENST00000539763.1 ENST00000587945.1 ENST00000536180.1 ENST00000589823.2 ENST00000592106.1 ENST00000591315.1 ENST00000545362.1 |
VMP1 |
vacuole membrane protein 1 |
| chr6_-_43484718 | 0.10 |
ENST00000372422.2 |
YIPF3 |
Yip1 domain family, member 3 |
| chr7_-_91509986 | 0.10 |
ENST00000456229.1 ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF |
mitochondrial transcription termination factor |
| chr2_+_201754135 | 0.10 |
ENST00000409357.1 ENST00000409129.2 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr10_-_18948156 | 0.10 |
ENST00000414939.1 ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1 |
ARL5B antisense RNA 1 |
| chr1_-_236767779 | 0.10 |
ENST00000366579.1 ENST00000366582.3 ENST00000366581.2 |
HEATR1 |
HEAT repeat containing 1 |
| chr1_+_156698234 | 0.10 |
ENST00000368218.4 ENST00000368216.4 |
RRNAD1 |
ribosomal RNA adenine dimethylase domain containing 1 |
| chr11_-_60929074 | 0.10 |
ENST00000301765.5 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
| chr17_+_4843654 | 0.10 |
ENST00000575111.1 |
RNF167 |
ring finger protein 167 |
| chr2_+_201754050 | 0.10 |
ENST00000426253.1 ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr1_+_11866270 | 0.10 |
ENST00000376497.3 ENST00000376487.3 ENST00000376496.3 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr8_-_144911469 | 0.10 |
ENST00000527744.1 ENST00000456095.2 ENST00000531897.1 ENST00000527197.1 ENST00000526459.1 ENST00000533162.1 ENST00000349157.6 |
PUF60 |
poly-U binding splicing factor 60KDa |
| chr1_-_11865982 | 0.10 |
ENST00000418034.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr8_-_95565673 | 0.10 |
ENST00000437199.1 ENST00000297591.5 ENST00000421249.2 |
KIAA1429 |
KIAA1429 |
| chr1_+_100316041 | 0.10 |
ENST00000370165.3 ENST00000370163.3 ENST00000294724.4 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr2_+_118572226 | 0.10 |
ENST00000263239.2 |
DDX18 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr17_+_4843413 | 0.10 |
ENST00000572430.1 ENST00000262482.6 |
RNF167 |
ring finger protein 167 |
| chr2_+_113885138 | 0.10 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr6_-_43484621 | 0.10 |
ENST00000506469.1 ENST00000503972.1 |
YIPF3 |
Yip1 domain family, member 3 |
| chr19_-_1174226 | 0.10 |
ENST00000587024.1 ENST00000361757.3 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
| chr16_+_8891670 | 0.10 |
ENST00000268261.4 ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2 |
phosphomannomutase 2 |
| chr7_-_108209897 | 0.10 |
ENST00000313516.5 |
THAP5 |
THAP domain containing 5 |
| chr1_-_155990580 | 0.10 |
ENST00000531917.1 ENST00000480567.1 ENST00000526212.1 ENST00000529008.1 ENST00000496742.1 ENST00000295702.4 |
SSR2 |
signal sequence receptor, beta (translocon-associated protein beta) |
| chr12_+_1800179 | 0.10 |
ENST00000357103.4 |
ADIPOR2 |
adiponectin receptor 2 |
| chrX_-_74376108 | 0.10 |
ENST00000339447.4 ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7 |
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr6_-_86303833 | 0.10 |
ENST00000505648.1 |
SNX14 |
sorting nexin 14 |
| chr14_-_78083112 | 0.10 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr11_+_60681346 | 0.10 |
ENST00000227525.3 |
TMEM109 |
transmembrane protein 109 |
| chr18_-_29522989 | 0.10 |
ENST00000582539.1 ENST00000283351.4 ENST00000582513.1 |
TRAPPC8 |
trafficking protein particle complex 8 |
| chr11_-_71791518 | 0.10 |
ENST00000537217.1 ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
| chr9_+_88556036 | 0.09 |
ENST00000361671.5 ENST00000416045.1 |
NAA35 |
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr8_-_144911181 | 0.09 |
ENST00000453551.2 ENST00000313352.7 ENST00000529999.1 |
PUF60 |
poly-U binding splicing factor 60KDa |
| chrY_-_21239221 | 0.09 |
ENST00000447937.1 ENST00000331787.2 |
TTTY14 |
testis-specific transcript, Y-linked 14 (non-protein coding) |
| chr18_+_20513278 | 0.09 |
ENST00000327155.5 |
RBBP8 |
retinoblastoma binding protein 8 |
| chr12_+_12966250 | 0.09 |
ENST00000352940.4 ENST00000358007.3 ENST00000544400.1 |
DDX47 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr22_-_38349552 | 0.09 |
ENST00000422191.1 ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23 |
chromosome 22 open reading frame 23 |
| chr9_-_35096545 | 0.09 |
ENST00000378617.3 ENST00000341666.3 ENST00000361778.2 |
PIGO |
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr18_+_33552597 | 0.09 |
ENST00000269194.6 ENST00000587873.1 |
C18orf21 |
chromosome 18 open reading frame 21 |
| chr10_-_14996070 | 0.09 |
ENST00000378258.1 ENST00000453695.2 ENST00000378246.2 |
DCLRE1C |
DNA cross-link repair 1C |
| chr6_+_111303218 | 0.09 |
ENST00000441448.2 |
RPF2 |
ribosome production factor 2 homolog (S. cerevisiae) |
| chr19_-_55791563 | 0.09 |
ENST00000588971.1 ENST00000255631.5 ENST00000587551.1 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr6_-_31620455 | 0.09 |
ENST00000437771.1 ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6 |
BCL2-associated athanogene 6 |
| chr16_-_1470709 | 0.09 |
ENST00000563974.1 ENST00000442039.2 |
C16orf91 |
chromosome 16 open reading frame 91 |
| chr14_-_21979428 | 0.09 |
ENST00000538267.1 ENST00000298717.4 |
METTL3 |
methyltransferase like 3 |
| chr17_+_4843679 | 0.09 |
ENST00000576229.1 |
RNF167 |
ring finger protein 167 |
| chr17_+_4981535 | 0.09 |
ENST00000318833.3 |
ZFP3 |
ZFP3 zinc finger protein |
| chr12_+_69080734 | 0.09 |
ENST00000378905.2 |
NUP107 |
nucleoporin 107kDa |
| chr5_+_10250328 | 0.09 |
ENST00000515390.1 |
CCT5 |
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr16_-_12009833 | 0.08 |
ENST00000420576.2 |
GSPT1 |
G1 to S phase transition 1 |
| chr19_-_11039261 | 0.08 |
ENST00000590329.1 ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2 |
Yip1 domain family, member 2 |
| chr5_-_132202329 | 0.08 |
ENST00000378673.2 |
GDF9 |
growth differentiation factor 9 |
| chr18_+_21033239 | 0.08 |
ENST00000581585.1 ENST00000577501.1 |
RIOK3 |
RIO kinase 3 |
| chr16_-_12009735 | 0.08 |
ENST00000439887.2 ENST00000434724.2 |
GSPT1 |
G1 to S phase transition 1 |
| chr11_-_125773085 | 0.08 |
ENST00000227474.3 ENST00000534158.1 ENST00000529801.1 |
PUS3 |
pseudouridylate synthase 3 |
| chr7_-_75115548 | 0.08 |
ENST00000453279.2 |
POM121C |
POM121 transmembrane nucleoporin C |
| chr15_+_34517251 | 0.08 |
ENST00000559421.1 |
EMC4 |
ER membrane protein complex subunit 4 |
| chr1_+_40506392 | 0.08 |
ENST00000414893.1 ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr22_-_30752861 | 0.08 |
ENST00000439242.1 ENST00000215793.8 |
SF3A1 |
splicing factor 3a, subunit 1, 120kDa |
| chr2_+_96931834 | 0.08 |
ENST00000488633.1 |
CIAO1 |
cytosolic iron-sulfur protein assembly 1 |
| chr16_-_4466565 | 0.08 |
ENST00000572467.1 ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16 CORO7 |
CORO7-PAM16 readthrough coronin 7 |
| chr7_-_1543981 | 0.08 |
ENST00000404767.3 |
INTS1 |
integrator complex subunit 1 |
| chr5_+_82373379 | 0.08 |
ENST00000396027.4 ENST00000511817.1 |
XRCC4 |
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr3_-_156272924 | 0.08 |
ENST00000467789.1 ENST00000265044.2 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr19_-_55791540 | 0.08 |
ENST00000433386.2 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr2_-_43823119 | 0.08 |
ENST00000403856.1 ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA |
thyroid adenoma associated |
| chr1_-_28415075 | 0.08 |
ENST00000373863.3 ENST00000436342.2 ENST00000540618.1 ENST00000545175.1 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
| chr2_+_9563697 | 0.08 |
ENST00000238112.3 |
CPSF3 |
cleavage and polyadenylation specific factor 3, 73kDa |
| chr10_+_127408263 | 0.08 |
ENST00000337623.3 |
C10orf137 |
erythroid differentiation regulatory factor 1 |
| chr13_+_46039037 | 0.08 |
ENST00000349995.5 |
COG3 |
component of oligomeric golgi complex 3 |
| chr18_+_20513782 | 0.08 |
ENST00000399722.2 ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8 |
retinoblastoma binding protein 8 |
| chr1_-_11866034 | 0.08 |
ENST00000376590.3 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr17_+_26662597 | 0.08 |
ENST00000544907.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr11_-_123612319 | 0.08 |
ENST00000526252.1 ENST00000530393.1 ENST00000533463.1 ENST00000336139.4 ENST00000529691.1 ENST00000528306.1 |
ZNF202 |
zinc finger protein 202 |
| chr5_+_82373317 | 0.08 |
ENST00000282268.3 ENST00000338635.6 |
XRCC4 |
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr1_+_155247207 | 0.07 |
ENST00000368358.3 |
HCN3 |
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr17_+_41561317 | 0.07 |
ENST00000540306.1 ENST00000262415.3 ENST00000605777.1 |
DHX8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr15_+_90234028 | 0.07 |
ENST00000268130.7 ENST00000560294.1 ENST00000558000.1 |
WDR93 |
WD repeat domain 93 |
| chr7_+_108210012 | 0.07 |
ENST00000249356.3 |
DNAJB9 |
DnaJ (Hsp40) homolog, subfamily B, member 9 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.3 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.3 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.2 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.2 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.1 | 0.2 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.3 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.0 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.0 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.3 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.7 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:2000843 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.0 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.0 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.0 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |