Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
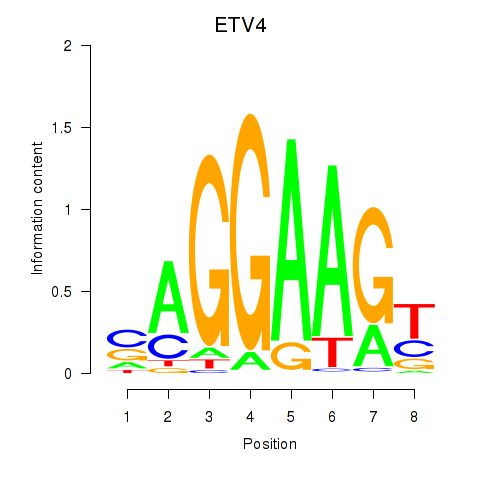
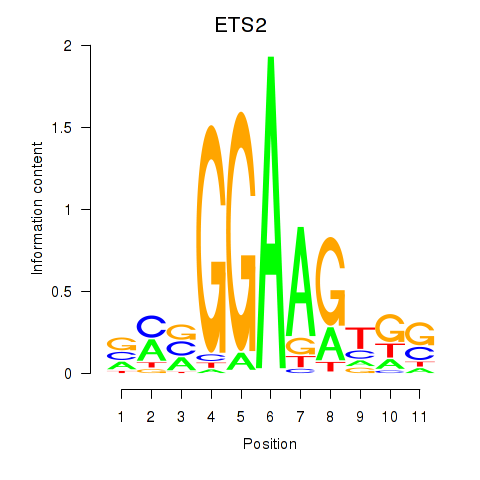
Results for ETV4_ETS2
Z-value: 0.43
Transcription factors associated with ETV4_ETS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV4
|
ENSG00000175832.8 | ETV4 |
|
ETS2
|
ENSG00000157557.7 | ETS2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV4 | hg19_v2_chr17_-_41623691_41623715, hg19_v2_chr17_-_41623075_41623101 | 0.53 | 1.8e-01 | Click! |
| ETS2 | hg19_v2_chr21_+_40177755_40177875, hg19_v2_chr21_+_40177143_40177231 | -0.21 | 6.2e-01 | Click! |
Activity profile of ETV4_ETS2 motif
Sorted Z-values of ETV4_ETS2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV4_ETS2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_62389449 | 0.22 |
ENST00000534026.1 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr18_+_21452964 | 0.19 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr18_+_21452804 | 0.17 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr11_+_63655987 | 0.13 |
ENST00000509502.2 ENST00000512060.1 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr10_-_98031310 | 0.12 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr2_+_37458928 | 0.12 |
ENST00000439218.1 ENST00000432075.1 |
NDUFAF7 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr5_+_149380256 | 0.12 |
ENST00000502717.1 |
HMGXB3 |
HMG box domain containing 3 |
| chr1_-_894620 | 0.11 |
ENST00000327044.6 |
NOC2L |
nucleolar complex associated 2 homolog (S. cerevisiae) |
| chr10_-_98031265 | 0.11 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr15_+_75074385 | 0.10 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr15_+_77287426 | 0.10 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr3_+_109128837 | 0.10 |
ENST00000497996.1 |
RP11-702L6.4 |
RP11-702L6.4 |
| chr17_-_56595196 | 0.10 |
ENST00000579921.1 ENST00000579925.1 ENST00000323456.5 |
MTMR4 |
myotubularin related protein 4 |
| chr19_-_49243845 | 0.10 |
ENST00000222145.4 |
RASIP1 |
Ras interacting protein 1 |
| chr6_-_11779403 | 0.10 |
ENST00000414691.3 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr3_-_172428959 | 0.10 |
ENST00000475381.1 ENST00000538775.1 ENST00000273512.3 ENST00000543711.1 |
NCEH1 |
neutral cholesterol ester hydrolase 1 |
| chr14_+_77924204 | 0.09 |
ENST00000555133.1 |
AHSA1 |
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr6_+_43739697 | 0.09 |
ENST00000230480.6 |
VEGFA |
vascular endothelial growth factor A |
| chr3_-_127842612 | 0.09 |
ENST00000417360.1 ENST00000322623.5 |
RUVBL1 |
RuvB-like AAA ATPase 1 |
| chr15_+_75074410 | 0.09 |
ENST00000439220.2 |
CSK |
c-src tyrosine kinase |
| chr10_+_102747783 | 0.09 |
ENST00000311916.2 ENST00000370228.1 |
C10orf2 |
chromosome 10 open reading frame 2 |
| chr1_-_52870104 | 0.08 |
ENST00000371568.3 |
ORC1 |
origin recognition complex, subunit 1 |
| chr1_+_153330322 | 0.08 |
ENST00000368738.3 |
S100A9 |
S100 calcium binding protein A9 |
| chr6_-_11779174 | 0.08 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr1_-_52870059 | 0.08 |
ENST00000371566.1 |
ORC1 |
origin recognition complex, subunit 1 |
| chr19_-_55791431 | 0.08 |
ENST00000593263.1 ENST00000376343.3 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr22_-_37880543 | 0.08 |
ENST00000442496.1 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_-_11779014 | 0.08 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr1_-_147232669 | 0.08 |
ENST00000369237.1 |
GJA5 |
gap junction protein, alpha 5, 40kDa |
| chr10_+_127408110 | 0.08 |
ENST00000356792.4 |
C10orf137 |
erythroid differentiation regulatory factor 1 |
| chr3_-_51975942 | 0.08 |
ENST00000232888.6 |
RRP9 |
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr5_+_179220979 | 0.08 |
ENST00000292596.10 ENST00000401985.3 |
LTC4S |
leukotriene C4 synthase |
| chr16_+_84682108 | 0.08 |
ENST00000564996.1 ENST00000258157.5 ENST00000567410.1 |
KLHL36 |
kelch-like family member 36 |
| chr14_+_77924373 | 0.08 |
ENST00000216479.3 ENST00000535854.2 ENST00000555517.1 |
AHSA1 |
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr9_-_33473882 | 0.07 |
ENST00000455041.2 ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6 |
nucleolar protein 6 (RNA-associated) |
| chr5_-_87564620 | 0.07 |
ENST00000506536.1 ENST00000512429.1 ENST00000514135.1 ENST00000296595.6 ENST00000509387.1 |
TMEM161B |
transmembrane protein 161B |
| chr19_-_11039188 | 0.07 |
ENST00000588347.1 |
YIPF2 |
Yip1 domain family, member 2 |
| chr11_+_131781290 | 0.07 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
| chr2_+_182321925 | 0.07 |
ENST00000339307.4 ENST00000397033.2 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
| chr2_+_234160217 | 0.07 |
ENST00000392017.4 ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
| chr1_-_200589859 | 0.07 |
ENST00000367350.4 |
KIF14 |
kinesin family member 14 |
| chr4_-_174255400 | 0.07 |
ENST00000506267.1 |
HMGB2 |
high mobility group box 2 |
| chr1_+_43766642 | 0.07 |
ENST00000372476.3 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr2_+_234160340 | 0.07 |
ENST00000417017.1 ENST00000392020.4 ENST00000392018.1 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
| chr5_-_176037105 | 0.07 |
ENST00000303991.4 |
GPRIN1 |
G protein regulated inducer of neurite outgrowth 1 |
| chr15_+_90744533 | 0.07 |
ENST00000411539.2 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr17_+_32646055 | 0.07 |
ENST00000394620.1 |
CCL8 |
chemokine (C-C motif) ligand 8 |
| chr8_-_134309335 | 0.07 |
ENST00000522890.1 ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1 |
N-myc downstream regulated 1 |
| chr1_+_186798073 | 0.07 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr16_+_29674540 | 0.06 |
ENST00000436527.1 ENST00000360121.3 ENST00000449759.1 |
SPN QPRT |
sialophorin quinolinate phosphoribosyltransferase |
| chr18_-_33077868 | 0.06 |
ENST00000590757.1 ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C RP11-322E11.6 |
INO80 complex subunit C Uncharacterized protein |
| chr8_-_104427313 | 0.06 |
ENST00000297578.4 |
SLC25A32 |
solute carrier family 25 (mitochondrial folate carrier), member 32 |
| chr3_-_112693865 | 0.06 |
ENST00000471858.1 ENST00000295863.4 ENST00000308611.3 |
CD200R1 |
CD200 receptor 1 |
| chr10_-_99161033 | 0.06 |
ENST00000315563.6 ENST00000370992.4 ENST00000414986.1 |
RRP12 |
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr5_+_180650271 | 0.06 |
ENST00000351937.5 ENST00000315073.5 |
TRIM41 |
tripartite motif containing 41 |
| chr7_-_25702669 | 0.06 |
ENST00000446840.1 |
AC003090.1 |
AC003090.1 |
| chr11_-_62389621 | 0.06 |
ENST00000531383.1 ENST00000265471.5 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr12_+_94071129 | 0.06 |
ENST00000552983.1 ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD |
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr10_-_43892668 | 0.06 |
ENST00000544000.1 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
| chr20_-_48532046 | 0.06 |
ENST00000543716.1 |
SPATA2 |
spermatogenesis associated 2 |
| chr5_+_82373317 | 0.06 |
ENST00000282268.3 ENST00000338635.6 |
XRCC4 |
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr1_-_201368707 | 0.06 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr5_+_149737202 | 0.06 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
| chr1_-_201368653 | 0.06 |
ENST00000367313.3 |
LAD1 |
ladinin 1 |
| chr12_+_94071341 | 0.06 |
ENST00000542893.2 |
CRADD |
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr1_-_236767779 | 0.06 |
ENST00000366579.1 ENST00000366582.3 ENST00000366581.2 |
HEATR1 |
HEAT repeat containing 1 |
| chr8_+_61591337 | 0.06 |
ENST00000423902.2 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
| chr19_-_55791563 | 0.06 |
ENST00000588971.1 ENST00000255631.5 ENST00000587551.1 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr5_+_179125368 | 0.06 |
ENST00000502296.1 ENST00000504734.1 ENST00000415618.2 |
CANX |
calnexin |
| chr6_-_155635583 | 0.06 |
ENST00000367166.4 |
TFB1M |
transcription factor B1, mitochondrial |
| chr15_-_41120896 | 0.06 |
ENST00000299174.5 ENST00000427255.2 |
PPP1R14D |
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr19_-_56632592 | 0.06 |
ENST00000587279.1 ENST00000270459.3 |
ZNF787 |
zinc finger protein 787 |
| chr6_+_31926857 | 0.06 |
ENST00000375394.2 ENST00000544581.1 |
SKIV2L |
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr2_-_235405168 | 0.06 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr5_+_82373379 | 0.06 |
ENST00000396027.4 ENST00000511817.1 |
XRCC4 |
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr21_+_26934165 | 0.06 |
ENST00000456917.1 |
MIR155HG |
MIR155 host gene (non-protein coding) |
| chr1_-_118472216 | 0.06 |
ENST00000369443.5 |
GDAP2 |
ganglioside induced differentiation associated protein 2 |
| chr2_+_103035102 | 0.06 |
ENST00000264260.2 |
IL18RAP |
interleukin 18 receptor accessory protein |
| chr1_+_6086354 | 0.06 |
ENST00000389632.4 ENST00000428161.1 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr19_-_55791058 | 0.06 |
ENST00000587959.1 ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr12_+_7079944 | 0.06 |
ENST00000261406.6 |
EMG1 |
EMG1 N1-specific pseudouridine methyltransferase |
| chr2_+_96931834 | 0.06 |
ENST00000488633.1 |
CIAO1 |
cytosolic iron-sulfur protein assembly 1 |
| chr14_-_24658053 | 0.06 |
ENST00000354464.6 |
IPO4 |
importin 4 |
| chr17_+_7942335 | 0.06 |
ENST00000380183.4 ENST00000572022.1 ENST00000380173.2 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr5_+_140430979 | 0.06 |
ENST00000306549.3 |
PCDHB1 |
protocadherin beta 1 |
| chr11_-_72432950 | 0.06 |
ENST00000426523.1 ENST00000429686.1 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr4_-_139163491 | 0.06 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr18_-_33077556 | 0.06 |
ENST00000589273.1 ENST00000586489.1 |
INO80C |
INO80 complex subunit C |
| chr13_-_25496926 | 0.06 |
ENST00000545981.1 ENST00000381884.4 |
CENPJ |
centromere protein J |
| chr19_-_55791540 | 0.06 |
ENST00000433386.2 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr3_+_38080691 | 0.06 |
ENST00000308059.6 ENST00000346219.3 ENST00000452631.2 |
DLEC1 |
deleted in lung and esophageal cancer 1 |
| chr17_+_7942424 | 0.06 |
ENST00000573359.1 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr3_+_119316721 | 0.06 |
ENST00000488919.1 ENST00000495992.1 |
PLA1A |
phospholipase A1 member A |
| chr3_+_47324424 | 0.06 |
ENST00000437353.1 ENST00000232766.5 ENST00000455924.2 |
KLHL18 |
kelch-like family member 18 |
| chr10_-_43892279 | 0.05 |
ENST00000443950.2 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
| chr11_-_9336234 | 0.05 |
ENST00000528080.1 |
TMEM41B |
transmembrane protein 41B |
| chr3_+_119316689 | 0.05 |
ENST00000273371.4 |
PLA1A |
phospholipase A1 member A |
| chr5_-_79287060 | 0.05 |
ENST00000512560.1 ENST00000509852.1 ENST00000512528.1 |
MTX3 |
metaxin 3 |
| chr16_+_57673430 | 0.05 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr2_+_32853093 | 0.05 |
ENST00000448773.1 ENST00000317907.4 |
TTC27 |
tetratricopeptide repeat domain 27 |
| chr12_+_107712173 | 0.05 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr4_-_682960 | 0.05 |
ENST00000512249.1 ENST00000515118.1 ENST00000347950.5 |
MFSD7 |
major facilitator superfamily domain containing 7 |
| chr4_+_110736659 | 0.05 |
ENST00000394631.3 ENST00000226796.6 |
GAR1 |
GAR1 ribonucleoprotein |
| chr1_-_183560011 | 0.05 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr8_+_121457642 | 0.05 |
ENST00000305949.1 |
MTBP |
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr16_-_2097787 | 0.05 |
ENST00000566380.1 ENST00000219066.1 |
NTHL1 |
nth endonuclease III-like 1 (E. coli) |
| chr3_-_112693759 | 0.05 |
ENST00000440122.2 ENST00000490004.1 |
CD200R1 |
CD200 receptor 1 |
| chr5_+_892745 | 0.05 |
ENST00000166345.3 |
TRIP13 |
thyroid hormone receptor interactor 13 |
| chr20_+_49126881 | 0.05 |
ENST00000371621.3 ENST00000541713.1 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
| chr19_-_4723761 | 0.05 |
ENST00000597849.1 ENST00000598800.1 ENST00000602161.1 ENST00000597726.1 ENST00000601130.1 ENST00000262960.9 |
DPP9 |
dipeptidyl-peptidase 9 |
| chr16_+_28505955 | 0.05 |
ENST00000564831.1 ENST00000328423.5 ENST00000431282.1 |
APOBR |
apolipoprotein B receptor |
| chr17_+_41561317 | 0.05 |
ENST00000540306.1 ENST00000262415.3 ENST00000605777.1 |
DHX8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr12_+_29302119 | 0.05 |
ENST00000536681.3 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr16_+_70557685 | 0.05 |
ENST00000302516.5 ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3 |
splicing factor 3b, subunit 3, 130kDa |
| chr11_-_118122996 | 0.05 |
ENST00000525386.1 ENST00000527472.1 ENST00000278949.4 |
MPZL3 |
myelin protein zero-like 3 |
| chr22_-_30752861 | 0.05 |
ENST00000439242.1 ENST00000215793.8 |
SF3A1 |
splicing factor 3a, subunit 1, 120kDa |
| chr2_-_69870747 | 0.05 |
ENST00000409068.1 |
AAK1 |
AP2 associated kinase 1 |
| chr15_+_41136586 | 0.05 |
ENST00000431806.1 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr9_+_116037922 | 0.05 |
ENST00000374198.4 |
PRPF4 |
pre-mRNA processing factor 4 |
| chr12_+_132195617 | 0.05 |
ENST00000261674.4 ENST00000535236.1 ENST00000541286.1 |
SFSWAP |
splicing factor, suppressor of white-apricot homolog (Drosophila) |
| chr9_+_37486005 | 0.05 |
ENST00000377792.3 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr11_-_64885111 | 0.05 |
ENST00000528598.1 ENST00000310597.4 |
ZNHIT2 |
zinc finger, HIT-type containing 2 |
| chr4_+_123844225 | 0.05 |
ENST00000274008.4 |
SPATA5 |
spermatogenesis associated 5 |
| chr11_+_63606373 | 0.05 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr5_+_179125907 | 0.05 |
ENST00000247461.4 ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX |
calnexin |
| chr21_-_34186006 | 0.05 |
ENST00000490358.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr11_+_2421718 | 0.05 |
ENST00000380996.5 ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4 |
tumor suppressing subtransferable candidate 4 |
| chr11_+_237016 | 0.05 |
ENST00000352303.5 |
PSMD13 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr11_+_63606477 | 0.05 |
ENST00000508192.1 ENST00000361128.5 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr19_-_4338783 | 0.05 |
ENST00000601482.1 ENST00000600324.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr19_-_17375527 | 0.05 |
ENST00000431146.2 ENST00000594190.1 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr6_-_31620455 | 0.05 |
ENST00000437771.1 ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6 |
BCL2-associated athanogene 6 |
| chr6_-_31620403 | 0.05 |
ENST00000451898.1 ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr3_+_148583043 | 0.05 |
ENST00000296046.3 |
CPA3 |
carboxypeptidase A3 (mast cell) |
| chr9_-_117150243 | 0.05 |
ENST00000374088.3 |
AKNA |
AT-hook transcription factor |
| chr3_-_47324242 | 0.04 |
ENST00000456548.1 ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9 |
kinesin family member 9 |
| chr5_-_132202329 | 0.04 |
ENST00000378673.2 |
GDF9 |
growth differentiation factor 9 |
| chr7_-_127032363 | 0.04 |
ENST00000393312.1 |
ZNF800 |
zinc finger protein 800 |
| chr19_-_4338838 | 0.04 |
ENST00000594605.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr6_-_31620149 | 0.04 |
ENST00000435080.1 ENST00000375976.4 ENST00000441054.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr1_-_111746966 | 0.04 |
ENST00000369752.5 |
DENND2D |
DENN/MADD domain containing 2D |
| chr5_-_58652788 | 0.04 |
ENST00000405755.2 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr12_-_108954933 | 0.04 |
ENST00000431469.2 ENST00000546815.1 |
SART3 |
squamous cell carcinoma antigen recognized by T cells 3 |
| chr11_+_236959 | 0.04 |
ENST00000431206.2 ENST00000528906.1 |
PSMD13 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr2_-_69870835 | 0.04 |
ENST00000409085.4 ENST00000406297.3 |
AAK1 |
AP2 associated kinase 1 |
| chr3_-_46249878 | 0.04 |
ENST00000296140.3 |
CCR1 |
chemokine (C-C motif) receptor 1 |
| chr11_+_236540 | 0.04 |
ENST00000532097.1 |
PSMD13 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr2_+_101869262 | 0.04 |
ENST00000289382.3 |
CNOT11 |
CCR4-NOT transcription complex, subunit 11 |
| chr21_-_33984888 | 0.04 |
ENST00000382549.4 ENST00000540881.1 |
C21orf59 |
chromosome 21 open reading frame 59 |
| chr1_-_205419053 | 0.04 |
ENST00000367154.1 |
LEMD1 |
LEM domain containing 1 |
| chr19_-_36004543 | 0.04 |
ENST00000339686.3 ENST00000447113.2 ENST00000440396.1 |
DMKN |
dermokine |
| chr10_-_96122682 | 0.04 |
ENST00000371361.3 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr17_-_7155274 | 0.04 |
ENST00000318988.6 ENST00000575783.1 ENST00000573600.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr16_+_67261008 | 0.04 |
ENST00000304800.9 ENST00000563953.1 ENST00000565201.1 |
TMEM208 |
transmembrane protein 208 |
| chr8_+_124194752 | 0.04 |
ENST00000318462.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr9_+_37485932 | 0.04 |
ENST00000377798.4 ENST00000442009.2 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr6_+_159290917 | 0.04 |
ENST00000367072.1 |
C6orf99 |
chromosome 6 open reading frame 99 |
| chr11_+_6260298 | 0.04 |
ENST00000379936.2 |
CNGA4 |
cyclic nucleotide gated channel alpha 4 |
| chr21_-_34185989 | 0.04 |
ENST00000487113.1 ENST00000382373.4 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr16_-_30125177 | 0.04 |
ENST00000406256.3 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr2_+_108994466 | 0.04 |
ENST00000272452.2 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr16_+_57653989 | 0.04 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr1_-_31769656 | 0.04 |
ENST00000446633.2 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr14_+_65171315 | 0.04 |
ENST00000394691.1 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr14_-_21979428 | 0.04 |
ENST00000538267.1 ENST00000298717.4 |
METTL3 |
methyltransferase like 3 |
| chr12_+_1800179 | 0.04 |
ENST00000357103.4 |
ADIPOR2 |
adiponectin receptor 2 |
| chr2_+_220462560 | 0.04 |
ENST00000456909.1 ENST00000295641.10 |
STK11IP |
serine/threonine kinase 11 interacting protein |
| chr11_-_47574664 | 0.04 |
ENST00000310513.5 ENST00000531165.1 |
CELF1 |
CUGBP, Elav-like family member 1 |
| chr5_+_61874562 | 0.04 |
ENST00000334994.5 ENST00000409534.1 |
LRRC70 IPO11 |
leucine rich repeat containing 70 importin 11 |
| chr11_+_34642656 | 0.04 |
ENST00000257831.3 ENST00000450654.2 |
EHF |
ets homologous factor |
| chr8_+_101170563 | 0.04 |
ENST00000520508.1 ENST00000388798.2 |
SPAG1 |
sperm associated antigen 1 |
| chr12_+_9102632 | 0.04 |
ENST00000539240.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr1_-_31769595 | 0.04 |
ENST00000263694.4 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr5_+_110427983 | 0.04 |
ENST00000513710.2 ENST00000505303.1 |
WDR36 |
WD repeat domain 36 |
| chr12_-_51785182 | 0.04 |
ENST00000356317.3 ENST00000603188.1 ENST00000604847.1 ENST00000604506.1 |
GALNT6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr11_+_65383227 | 0.04 |
ENST00000355703.3 |
PCNXL3 |
pecanex-like 3 (Drosophila) |
| chr17_-_42092313 | 0.04 |
ENST00000587529.1 ENST00000206380.3 ENST00000542039.1 |
TMEM101 |
transmembrane protein 101 |
| chr11_+_60467047 | 0.04 |
ENST00000300226.2 |
MS4A8 |
membrane-spanning 4-domains, subfamily A, member 8 |
| chr2_-_24583314 | 0.04 |
ENST00000443927.1 ENST00000406921.3 ENST00000412011.1 |
ITSN2 |
intersectin 2 |
| chr8_+_124194875 | 0.04 |
ENST00000522648.1 ENST00000276699.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr16_+_57653854 | 0.04 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr13_-_46626847 | 0.04 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr2_+_177134201 | 0.04 |
ENST00000452865.1 |
MTX2 |
metaxin 2 |
| chr19_-_12792020 | 0.04 |
ENST00000594424.1 ENST00000597152.1 ENST00000596162.1 |
DHPS |
deoxyhypusine synthase |
| chr3_-_47324008 | 0.04 |
ENST00000425853.1 |
KIF9 |
kinesin family member 9 |
| chr17_+_76126842 | 0.04 |
ENST00000590426.1 ENST00000590799.1 ENST00000318430.5 ENST00000589691.1 |
TMC8 |
transmembrane channel-like 8 |
| chr21_-_33984865 | 0.04 |
ENST00000458138.1 |
C21orf59 |
chromosome 21 open reading frame 59 |
| chr18_-_33077942 | 0.04 |
ENST00000334598.7 |
INO80C |
INO80 complex subunit C |
| chr22_+_39898325 | 0.04 |
ENST00000325301.2 ENST00000404569.1 |
MIEF1 |
mitochondrial elongation factor 1 |
| chr14_-_21924209 | 0.04 |
ENST00000557364.1 |
CHD8 |
chromodomain helicase DNA binding protein 8 |
| chr4_+_81256871 | 0.04 |
ENST00000358105.3 ENST00000508675.1 |
C4orf22 |
chromosome 4 open reading frame 22 |
| chr1_+_174417095 | 0.04 |
ENST00000367685.2 |
GPR52 |
G protein-coupled receptor 52 |
| chr19_-_59070239 | 0.04 |
ENST00000595957.1 ENST00000253023.3 |
UBE2M |
ubiquitin-conjugating enzyme E2M |
| chr10_+_75504105 | 0.04 |
ENST00000535742.1 ENST00000546025.1 ENST00000345254.4 ENST00000540668.1 ENST00000339365.2 ENST00000411652.2 |
SEC24C |
SEC24 family member C |
| chr1_+_203595903 | 0.04 |
ENST00000367218.3 ENST00000367219.3 ENST00000391954.2 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr19_+_35739897 | 0.04 |
ENST00000605618.1 ENST00000427250.1 ENST00000601623.1 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr1_+_955448 | 0.04 |
ENST00000379370.2 |
AGRN |
agrin |
| chr1_+_203595689 | 0.04 |
ENST00000357681.5 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr16_+_69373323 | 0.04 |
ENST00000254940.5 |
NIP7 |
NIP7, nucleolar pre-rRNA processing protein |
| chr17_-_76123101 | 0.04 |
ENST00000392467.3 |
TMC6 |
transmembrane channel-like 6 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0090313 | regulation of protein targeting to membrane(GO:0090313) positive regulation of protein targeting to membrane(GO:0090314) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.1 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.0 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:0035711 | T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.0 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.0 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.0 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.0 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.1 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |