Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
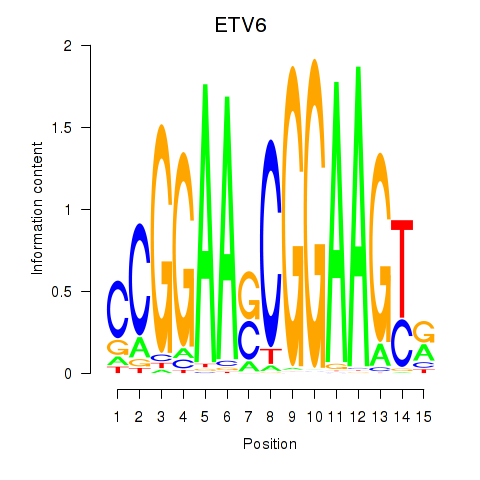
Results for ETV6
Z-value: 0.71
Transcription factors associated with ETV6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV6
|
ENSG00000139083.6 | ETV6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV6 | hg19_v2_chr12_+_11802753_11802834 | -0.94 | 5.3e-04 | Click! |
Activity profile of ETV6 motif
Sorted Z-values of ETV6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_225907150 | 0.81 |
ENST00000258390.7 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr12_+_120105558 | 0.37 |
ENST00000229328.5 ENST00000541640.1 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr13_-_29292956 | 0.31 |
ENST00000266943.6 |
SLC46A3 |
solute carrier family 46, member 3 |
| chr6_+_30524663 | 0.30 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr19_-_8942962 | 0.29 |
ENST00000601372.1 |
ZNF558 |
zinc finger protein 558 |
| chr1_+_22351977 | 0.29 |
ENST00000420503.1 ENST00000416769.1 ENST00000404210.2 |
LINC00339 |
long intergenic non-protein coding RNA 339 |
| chr16_+_3355472 | 0.29 |
ENST00000574298.1 |
ZNF75A |
zinc finger protein 75a |
| chr3_-_16555150 | 0.29 |
ENST00000334133.4 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr6_+_30525051 | 0.27 |
ENST00000376557.3 |
PRR3 |
proline rich 3 |
| chr16_-_3355645 | 0.26 |
ENST00000396862.1 ENST00000573608.1 |
TIGD7 |
tigger transposable element derived 7 |
| chr16_+_75256507 | 0.26 |
ENST00000495583.1 |
CTRB1 |
chymotrypsinogen B1 |
| chr6_-_31697255 | 0.25 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr1_-_149900122 | 0.24 |
ENST00000271628.8 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
| chrX_-_107018969 | 0.24 |
ENST00000372383.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr6_+_31620191 | 0.24 |
ENST00000375918.2 ENST00000375920.4 |
APOM |
apolipoprotein M |
| chr2_+_65454926 | 0.23 |
ENST00000542850.1 ENST00000377982.4 |
ACTR2 |
ARP2 actin-related protein 2 homolog (yeast) |
| chr4_+_6576895 | 0.22 |
ENST00000285599.3 ENST00000504248.1 ENST00000505907.1 |
MAN2B2 |
mannosidase, alpha, class 2B, member 2 |
| chr2_+_65454863 | 0.20 |
ENST00000260641.5 |
ACTR2 |
ARP2 actin-related protein 2 homolog (yeast) |
| chr3_-_179169330 | 0.20 |
ENST00000232564.3 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr6_-_31697563 | 0.19 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr16_+_30406721 | 0.19 |
ENST00000320159.2 |
ZNF48 |
zinc finger protein 48 |
| chr5_-_36152031 | 0.19 |
ENST00000296603.4 |
LMBRD2 |
LMBR1 domain containing 2 |
| chr19_+_1249869 | 0.18 |
ENST00000591446.2 |
MIDN |
midnolin |
| chr19_-_53606604 | 0.18 |
ENST00000599056.1 ENST00000599247.1 ENST00000355147.5 ENST00000429604.1 ENST00000418871.1 ENST00000599637.1 |
ZNF160 |
zinc finger protein 160 |
| chr5_-_150473127 | 0.18 |
ENST00000521001.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr17_+_75137460 | 0.18 |
ENST00000587820.1 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr12_-_58329819 | 0.18 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr7_-_112635675 | 0.18 |
ENST00000447785.1 ENST00000451962.1 |
AC018464.3 |
AC018464.3 |
| chr20_+_5931497 | 0.17 |
ENST00000378886.2 ENST00000265187.4 |
MCM8 |
minichromosome maintenance complex component 8 |
| chr6_-_31697977 | 0.17 |
ENST00000375787.2 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr16_-_3493528 | 0.17 |
ENST00000301744.4 |
ZNF597 |
zinc finger protein 597 |
| chrX_+_47092314 | 0.17 |
ENST00000218348.3 |
USP11 |
ubiquitin specific peptidase 11 |
| chr20_+_5931275 | 0.17 |
ENST00000378896.3 ENST00000378883.1 |
MCM8 |
minichromosome maintenance complex component 8 |
| chr2_+_152266604 | 0.17 |
ENST00000430328.2 |
RIF1 |
RAP1 interacting factor homolog (yeast) |
| chr6_-_11382478 | 0.17 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr4_+_153457404 | 0.17 |
ENST00000604157.1 ENST00000594836.1 |
MIR4453 |
microRNA 4453 |
| chr5_+_102455968 | 0.17 |
ENST00000358359.3 |
PPIP5K2 |
diphosphoinositol pentakisphosphate kinase 2 |
| chr19_-_9546177 | 0.17 |
ENST00000592292.1 ENST00000588221.1 |
ZNF266 |
zinc finger protein 266 |
| chr20_+_43595115 | 0.16 |
ENST00000372806.3 ENST00000396731.4 ENST00000372801.1 ENST00000499879.2 |
STK4 |
serine/threonine kinase 4 |
| chr19_-_31840438 | 0.16 |
ENST00000240587.4 |
TSHZ3 |
teashirt zinc finger homeobox 3 |
| chr5_+_102201430 | 0.16 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr9_-_72374848 | 0.16 |
ENST00000377200.5 ENST00000340434.4 ENST00000472967.2 |
PTAR1 |
protein prenyltransferase alpha subunit repeat containing 1 |
| chrX_+_46306624 | 0.16 |
ENST00000360017.5 |
KRBOX4 |
KRAB box domain containing 4 |
| chr3_-_123680246 | 0.16 |
ENST00000488653.2 |
CCDC14 |
coiled-coil domain containing 14 |
| chr2_+_189156721 | 0.16 |
ENST00000409927.1 ENST00000409805.1 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr20_+_39969519 | 0.16 |
ENST00000373257.3 |
LPIN3 |
lipin 3 |
| chr5_+_102201509 | 0.16 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr4_+_148538517 | 0.16 |
ENST00000296582.3 ENST00000508208.1 |
TMEM184C |
transmembrane protein 184C |
| chr2_+_189156586 | 0.15 |
ENST00000409830.1 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr20_-_33872518 | 0.15 |
ENST00000374436.3 |
EIF6 |
eukaryotic translation initiation factor 6 |
| chr9_-_14693417 | 0.15 |
ENST00000380916.4 |
ZDHHC21 |
zinc finger, DHHC-type containing 21 |
| chr9_-_80263220 | 0.15 |
ENST00000341700.6 |
GNA14 |
guanine nucleotide binding protein (G protein), alpha 14 |
| chrX_+_69509927 | 0.15 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr2_+_189156389 | 0.15 |
ENST00000409843.1 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr5_+_167913450 | 0.15 |
ENST00000231572.3 ENST00000538719.1 |
RARS |
arginyl-tRNA synthetase |
| chr19_-_9546227 | 0.15 |
ENST00000361451.2 ENST00000361151.1 |
ZNF266 |
zinc finger protein 266 |
| chr20_-_33872548 | 0.15 |
ENST00000374443.3 |
EIF6 |
eukaryotic translation initiation factor 6 |
| chr10_+_104503727 | 0.15 |
ENST00000448841.1 |
WBP1L |
WW domain binding protein 1-like |
| chr3_+_42977846 | 0.15 |
ENST00000383748.4 |
KRBOX1 |
KRAB box domain containing 1 |
| chr5_+_102201722 | 0.15 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr20_-_2821271 | 0.15 |
ENST00000448755.1 ENST00000360652.2 |
PCED1A |
PC-esterase domain containing 1A |
| chr2_+_219537015 | 0.14 |
ENST00000440309.1 ENST00000424080.1 |
STK36 |
serine/threonine kinase 36 |
| chr2_+_219536749 | 0.14 |
ENST00000295709.3 ENST00000392106.2 ENST00000392105.3 ENST00000455724.1 |
STK36 |
serine/threonine kinase 36 |
| chr17_+_26989109 | 0.14 |
ENST00000314616.6 ENST00000347486.4 |
SUPT6H |
suppressor of Ty 6 homolog (S. cerevisiae) |
| chr19_-_44860820 | 0.14 |
ENST00000354340.4 ENST00000337401.4 ENST00000587909.1 |
ZNF112 |
zinc finger protein 112 |
| chr3_-_122134882 | 0.14 |
ENST00000330689.4 |
WDR5B |
WD repeat domain 5B |
| chr5_+_102201687 | 0.14 |
ENST00000304400.7 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr12_+_56511943 | 0.14 |
ENST00000257940.2 ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10 ESYT1 |
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr20_-_3140490 | 0.14 |
ENST00000449731.1 ENST00000380266.3 |
UBOX5 FASTKD5 |
U-box domain containing 5 FAST kinase domains 5 |
| chr17_+_80186273 | 0.14 |
ENST00000584689.1 ENST00000392341.1 ENST00000583237.1 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr17_+_61904766 | 0.14 |
ENST00000581842.1 ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5 |
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr11_-_47600320 | 0.14 |
ENST00000525720.1 ENST00000531067.1 ENST00000533290.1 ENST00000529499.1 ENST00000529946.1 ENST00000526005.1 ENST00000395288.2 ENST00000534239.1 |
KBTBD4 |
kelch repeat and BTB (POZ) domain containing 4 |
| chr13_+_113623509 | 0.14 |
ENST00000535094.2 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr11_+_124824000 | 0.13 |
ENST00000529051.1 ENST00000344762.5 |
CCDC15 |
coiled-coil domain containing 15 |
| chr4_-_2366525 | 0.13 |
ENST00000515169.1 ENST00000515312.1 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
| chr15_-_51058005 | 0.13 |
ENST00000261854.5 |
SPPL2A |
signal peptide peptidase like 2A |
| chr19_+_57078854 | 0.13 |
ENST00000330619.8 ENST00000391709.3 ENST00000601902.1 |
ZNF470 |
zinc finger protein 470 |
| chr2_+_201981527 | 0.13 |
ENST00000441224.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr5_+_102455853 | 0.13 |
ENST00000515845.1 ENST00000321521.9 ENST00000507921.1 |
PPIP5K2 |
diphosphoinositol pentakisphosphate kinase 2 |
| chr16_-_31085514 | 0.13 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr9_-_125693757 | 0.12 |
ENST00000373656.3 |
ZBTB26 |
zinc finger and BTB domain containing 26 |
| chr19_+_9473676 | 0.12 |
ENST00000343499.4 ENST00000592912.1 |
ZNF177 |
zinc finger protein 177 |
| chr17_-_79876010 | 0.12 |
ENST00000328666.6 |
SIRT7 |
sirtuin 7 |
| chr16_+_77225071 | 0.12 |
ENST00000439557.2 ENST00000545553.1 |
MON1B |
MON1 secretory trafficking family member B |
| chr2_+_38152462 | 0.12 |
ENST00000354545.2 |
RMDN2 |
regulator of microtubule dynamics 2 |
| chr11_-_47600549 | 0.12 |
ENST00000430070.2 |
KBTBD4 |
kelch repeat and BTB (POZ) domain containing 4 |
| chr5_+_179247759 | 0.12 |
ENST00000389805.4 ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1 |
sequestosome 1 |
| chr17_-_38574169 | 0.12 |
ENST00000423485.1 |
TOP2A |
topoisomerase (DNA) II alpha 170kDa |
| chr12_+_124196865 | 0.12 |
ENST00000330342.3 |
ATP6V0A2 |
ATPase, H+ transporting, lysosomal V0 subunit a2 |
| chr17_+_18601299 | 0.12 |
ENST00000572555.1 ENST00000395902.3 ENST00000449552.2 |
TRIM16L |
tripartite motif containing 16-like |
| chr19_+_7069426 | 0.12 |
ENST00000252840.6 ENST00000414706.1 |
ZNF557 |
zinc finger protein 557 |
| chr20_-_5931051 | 0.11 |
ENST00000453074.2 |
TRMT6 |
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr16_-_28937027 | 0.11 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chrX_+_51546103 | 0.11 |
ENST00000375772.3 |
MAGED1 |
melanoma antigen family D, 1 |
| chr7_+_155090271 | 0.11 |
ENST00000476756.1 |
INSIG1 |
insulin induced gene 1 |
| chr2_+_128848881 | 0.11 |
ENST00000259253.6 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr1_-_114301755 | 0.11 |
ENST00000393357.2 ENST00000369596.2 ENST00000446739.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr11_+_58910201 | 0.11 |
ENST00000528737.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr1_-_32210275 | 0.11 |
ENST00000440175.2 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
| chrX_+_47342970 | 0.11 |
ENST00000357412.1 |
CXorf24 |
chromosome X open reading frame 24 |
| chr20_+_33464238 | 0.11 |
ENST00000360596.2 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
| chr14_-_93673353 | 0.11 |
ENST00000556566.1 ENST00000306954.4 |
C14orf142 |
chromosome 14 open reading frame 142 |
| chr14_+_55518349 | 0.11 |
ENST00000395468.4 |
MAPK1IP1L |
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr15_-_43559055 | 0.11 |
ENST00000220420.5 ENST00000349114.4 |
TGM5 |
transglutaminase 5 |
| chrX_+_11129388 | 0.11 |
ENST00000321143.4 ENST00000380763.3 ENST00000380762.4 |
HCCS |
holocytochrome c synthase |
| chr11_+_12399071 | 0.11 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr9_+_131267052 | 0.11 |
ENST00000539582.1 |
GLE1 |
GLE1 RNA export mediator |
| chr2_-_219537134 | 0.10 |
ENST00000295704.2 |
RNF25 |
ring finger protein 25 |
| chr5_-_137090028 | 0.10 |
ENST00000314940.4 |
HNRNPA0 |
heterogeneous nuclear ribonucleoprotein A0 |
| chr12_-_58329888 | 0.10 |
ENST00000546580.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr2_+_3383439 | 0.10 |
ENST00000382110.2 ENST00000324266.5 |
TRAPPC12 |
trafficking protein particle complex 12 |
| chr9_+_34458771 | 0.10 |
ENST00000437363.1 ENST00000242317.4 |
DNAI1 |
dynein, axonemal, intermediate chain 1 |
| chr3_+_40518599 | 0.10 |
ENST00000314686.5 ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619 |
zinc finger protein 619 |
| chr20_+_33464407 | 0.10 |
ENST00000253382.5 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
| chrX_+_110924346 | 0.10 |
ENST00000371979.3 ENST00000251943.4 ENST00000486353.1 ENST00000394780.3 ENST00000495283.1 |
ALG13 |
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr2_+_201981119 | 0.10 |
ENST00000395148.2 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr12_+_57522258 | 0.10 |
ENST00000553277.1 ENST00000243077.3 |
LRP1 |
low density lipoprotein receptor-related protein 1 |
| chr2_+_128848740 | 0.10 |
ENST00000375990.3 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr8_+_104427581 | 0.10 |
ENST00000521716.1 ENST00000521971.1 ENST00000519682.1 |
DCAF13 |
DDB1 and CUL4 associated factor 13 |
| chr11_+_58910295 | 0.10 |
ENST00000420244.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr9_-_115480303 | 0.10 |
ENST00000374234.1 ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP |
INTS3 and NABP interacting protein |
| chr2_+_64068844 | 0.10 |
ENST00000337130.5 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
| chr9_-_21335240 | 0.10 |
ENST00000537938.1 |
KLHL9 |
kelch-like family member 9 |
| chr20_-_5931109 | 0.10 |
ENST00000203001.2 |
TRMT6 |
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr2_+_64069459 | 0.10 |
ENST00000445915.2 ENST00000475462.1 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
| chr20_-_2821756 | 0.10 |
ENST00000356872.3 ENST00000439542.1 |
PCED1A |
PC-esterase domain containing 1A |
| chr9_-_86571628 | 0.10 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr19_+_49375649 | 0.09 |
ENST00000200453.5 |
PPP1R15A |
protein phosphatase 1, regulatory subunit 15A |
| chr19_+_50919056 | 0.09 |
ENST00000599632.1 |
CTD-2545M3.6 |
CTD-2545M3.6 |
| chr20_+_2821340 | 0.09 |
ENST00000380445.3 ENST00000380469.3 |
VPS16 |
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr19_+_14017116 | 0.09 |
ENST00000589606.1 |
CC2D1A |
coiled-coil and C2 domain containing 1A |
| chr1_-_165738072 | 0.09 |
ENST00000481278.1 |
TMCO1 |
transmembrane and coiled-coil domains 1 |
| chr17_+_79935464 | 0.09 |
ENST00000581647.1 ENST00000580534.1 ENST00000579684.1 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr21_-_46340807 | 0.09 |
ENST00000397846.3 ENST00000524251.1 ENST00000522688.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr19_-_45004556 | 0.09 |
ENST00000587047.1 ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180 |
zinc finger protein 180 |
| chr6_-_149969829 | 0.09 |
ENST00000367411.2 |
KATNA1 |
katanin p60 (ATPase containing) subunit A 1 |
| chr11_+_65479702 | 0.09 |
ENST00000530446.1 ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5 |
K(lysine) acetyltransferase 5 |
| chr16_-_15736881 | 0.09 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr9_-_33264557 | 0.09 |
ENST00000473781.1 ENST00000488499.1 |
BAG1 |
BCL2-associated athanogene |
| chr1_-_19811996 | 0.09 |
ENST00000264203.3 ENST00000401084.2 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
| chr19_-_40562063 | 0.09 |
ENST00000598845.1 ENST00000593605.1 ENST00000221355.6 ENST00000434248.1 |
ZNF780B |
zinc finger protein 780B |
| chr19_-_44952635 | 0.09 |
ENST00000592308.1 ENST00000588931.1 ENST00000291187.4 |
ZNF229 |
zinc finger protein 229 |
| chr1_+_43855560 | 0.09 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr7_+_39773160 | 0.09 |
ENST00000340510.4 |
LINC00265 |
long intergenic non-protein coding RNA 265 |
| chr17_+_79935418 | 0.09 |
ENST00000306729.7 ENST00000306739.4 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr16_-_15736953 | 0.09 |
ENST00000548025.1 ENST00000551742.1 ENST00000602337.1 ENST00000344181.3 ENST00000396368.3 |
KIAA0430 |
KIAA0430 |
| chr3_-_52008016 | 0.09 |
ENST00000489595.2 ENST00000461108.1 ENST00000395008.2 ENST00000525795.1 ENST00000361143.5 |
RP11-155D18.14 ABHD14B |
Poly(rC)-binding protein 4 abhydrolase domain containing 14B |
| chr4_-_159592996 | 0.09 |
ENST00000508457.1 |
C4orf46 |
chromosome 4 open reading frame 46 |
| chr19_-_1876156 | 0.09 |
ENST00000565797.1 |
CTB-31O20.2 |
CTB-31O20.2 |
| chr4_-_83812248 | 0.09 |
ENST00000514326.1 ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr4_-_159593179 | 0.09 |
ENST00000379205.4 |
C4orf46 |
chromosome 4 open reading frame 46 |
| chr7_+_44084233 | 0.09 |
ENST00000448521.1 |
DBNL |
drebrin-like |
| chr5_-_40755987 | 0.09 |
ENST00000337702.4 |
TTC33 |
tetratricopeptide repeat domain 33 |
| chr2_-_3595547 | 0.08 |
ENST00000438485.1 |
RP13-512J5.1 |
Uncharacterized protein |
| chr17_-_5389477 | 0.08 |
ENST00000572834.1 ENST00000570848.1 ENST00000571971.1 ENST00000158771.4 |
DERL2 |
derlin 2 |
| chr4_-_83812402 | 0.08 |
ENST00000395310.2 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr12_+_6982725 | 0.08 |
ENST00000433346.1 |
LRRC23 |
leucine rich repeat containing 23 |
| chr11_-_8190534 | 0.08 |
ENST00000309737.6 ENST00000425599.2 ENST00000539720.1 ENST00000531450.1 ENST00000419822.2 ENST00000335425.7 ENST00000343202.4 |
RIC3 |
RIC3 acetylcholine receptor chaperone |
| chr6_-_149969871 | 0.08 |
ENST00000335643.8 ENST00000444282.1 |
KATNA1 |
katanin p60 (ATPase containing) subunit A 1 |
| chr1_-_95285764 | 0.08 |
ENST00000414374.1 ENST00000421997.1 ENST00000418366.2 ENST00000452922.1 |
LINC01057 |
long intergenic non-protein coding RNA 1057 |
| chr7_+_100482595 | 0.08 |
ENST00000448764.1 |
SRRT |
serrate RNA effector molecule homolog (Arabidopsis) |
| chr7_-_229557 | 0.08 |
ENST00000514988.1 |
AC145676.2 |
Uncharacterized protein |
| chrX_-_100872911 | 0.08 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr8_-_77912431 | 0.08 |
ENST00000357039.4 ENST00000522527.1 |
PEX2 |
peroxisomal biogenesis factor 2 |
| chr2_-_85839146 | 0.08 |
ENST00000306336.5 ENST00000409734.3 |
C2orf68 |
chromosome 2 open reading frame 68 |
| chr6_+_170102210 | 0.08 |
ENST00000439249.1 ENST00000332290.2 |
C6orf120 |
chromosome 6 open reading frame 120 |
| chr13_+_96329381 | 0.08 |
ENST00000602402.1 ENST00000376795.6 |
DNAJC3 |
DnaJ (Hsp40) homolog, subfamily C, member 3 |
| chr11_+_43380459 | 0.08 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chr5_-_40798473 | 0.08 |
ENST00000354209.3 |
PRKAA1 |
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr10_-_79789291 | 0.08 |
ENST00000372371.3 |
POLR3A |
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr19_+_14017003 | 0.08 |
ENST00000318003.7 |
CC2D1A |
coiled-coil and C2 domain containing 1A |
| chr17_+_7487146 | 0.08 |
ENST00000396501.4 ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1 |
mannose-P-dolichol utilization defect 1 |
| chr17_-_37844267 | 0.08 |
ENST00000579146.1 ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3 |
post-GPI attachment to proteins 3 |
| chr6_-_52926539 | 0.08 |
ENST00000350082.5 ENST00000356971.3 |
ICK |
intestinal cell (MAK-like) kinase |
| chr19_-_16770915 | 0.08 |
ENST00000593459.1 ENST00000358726.6 ENST00000597711.1 ENST00000487416.2 |
CTC-429P9.4 SMIM7 |
Small integral membrane protein 7; Uncharacterized protein small integral membrane protein 7 |
| chr19_-_6393216 | 0.08 |
ENST00000595047.1 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
| chr15_+_41624892 | 0.08 |
ENST00000260359.6 ENST00000450318.1 ENST00000450592.2 ENST00000559596.1 ENST00000414849.2 ENST00000560747.1 ENST00000560177.1 |
NUSAP1 |
nucleolar and spindle associated protein 1 |
| chr19_-_9929708 | 0.08 |
ENST00000247977.4 ENST00000590277.1 ENST00000588922.1 ENST00000589626.1 ENST00000592067.1 ENST00000586469.1 |
FBXL12 |
F-box and leucine-rich repeat protein 12 |
| chr17_-_74099772 | 0.08 |
ENST00000411744.2 ENST00000332065.5 |
EXOC7 |
exocyst complex component 7 |
| chr10_+_26986582 | 0.08 |
ENST00000376215.5 ENST00000376203.5 |
PDSS1 |
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr4_+_1723197 | 0.07 |
ENST00000485989.2 ENST00000313288.4 |
TACC3 |
transforming, acidic coiled-coil containing protein 3 |
| chr10_-_69835099 | 0.07 |
ENST00000373700.4 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chrX_-_47863348 | 0.07 |
ENST00000376943.3 ENST00000396965.1 ENST00000305127.6 |
ZNF182 |
zinc finger protein 182 |
| chr11_+_2415061 | 0.07 |
ENST00000481687.1 |
CD81 |
CD81 molecule |
| chr5_-_443239 | 0.07 |
ENST00000408966.2 |
C5orf55 |
chromosome 5 open reading frame 55 |
| chr2_+_122494676 | 0.07 |
ENST00000455432.1 |
TSN |
translin |
| chr4_+_8271471 | 0.07 |
ENST00000307358.2 ENST00000382512.3 |
HTRA3 |
HtrA serine peptidase 3 |
| chr12_-_56223394 | 0.07 |
ENST00000317269.3 |
DNAJC14 |
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr5_+_73109339 | 0.07 |
ENST00000296799.4 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr19_-_37263693 | 0.07 |
ENST00000591344.1 |
ZNF850 |
zinc finger protein 850 |
| chr22_-_19166343 | 0.07 |
ENST00000215882.5 |
SLC25A1 |
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chrX_+_46404928 | 0.07 |
ENST00000421685.2 ENST00000609887.1 |
ZNF674-AS1 |
ZNF674 antisense RNA 1 (head to head) |
| chr10_+_37414785 | 0.07 |
ENST00000374660.1 ENST00000602533.1 ENST00000361713.1 |
ANKRD30A |
ankyrin repeat domain 30A |
| chr17_-_74099795 | 0.07 |
ENST00000406660.3 ENST00000335146.7 ENST00000405575.4 ENST00000589210.1 ENST00000607838.1 |
EXOC7 |
exocyst complex component 7 |
| chr14_+_45431379 | 0.07 |
ENST00000361577.3 ENST00000361462.2 ENST00000382233.2 |
FAM179B |
family with sequence similarity 179, member B |
| chr17_+_19030782 | 0.07 |
ENST00000344415.4 ENST00000577213.1 |
GRAPL |
GRB2-related adaptor protein-like |
| chr9_+_72873837 | 0.07 |
ENST00000361138.5 |
SMC5 |
structural maintenance of chromosomes 5 |
| chr22_+_40742512 | 0.07 |
ENST00000454266.2 ENST00000342312.6 |
ADSL |
adenylosuccinate lyase |
| chr8_-_48872686 | 0.07 |
ENST00000314191.2 ENST00000338368.3 |
PRKDC |
protein kinase, DNA-activated, catalytic polypeptide |
| chr16_+_66638003 | 0.07 |
ENST00000562357.1 ENST00000360086.4 ENST00000562707.1 ENST00000361909.4 ENST00000460097.1 ENST00000565666.1 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.4 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.2 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 0.2 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.3 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.2 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.2 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.2 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.6 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.4 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.0 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.0 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0048733 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.2 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0042954 | apolipoprotein receptor activity(GO:0030226) lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.0 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |