Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for FOXA2_FOXJ3
Z-value: 0.83
Transcription factors associated with FOXA2_FOXJ3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
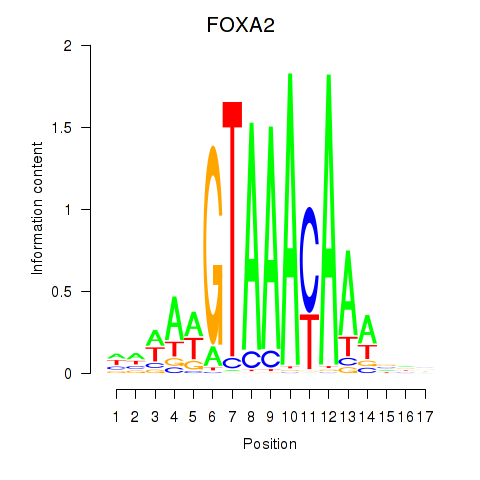
FOXA2
|
ENSG00000125798.10 | FOXA2 |
|
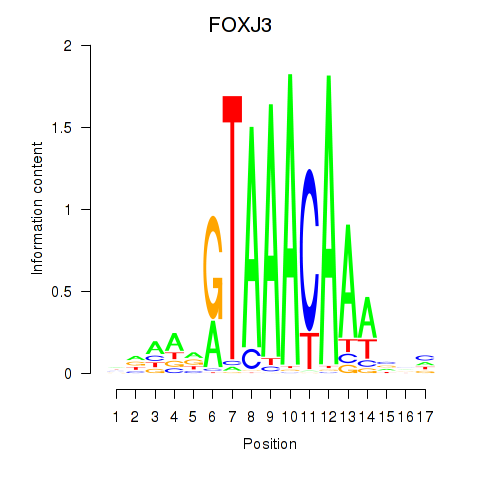
FOXJ3
|
ENSG00000198815.4 | FOXJ3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA2 | hg19_v2_chr20_-_22566089_22566097, hg19_v2_chr20_-_22565101_22565223 | -0.87 | 4.5e-03 | Click! |
| FOXJ3 | hg19_v2_chr1_-_42800614_42800649, hg19_v2_chr1_-_42801540_42801562, hg19_v2_chr1_-_42800860_42800912 | -0.82 | 1.2e-02 | Click! |
Activity profile of FOXA2_FOXJ3 motif
Sorted Z-values of FOXA2_FOXJ3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA2_FOXJ3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91574142 | 2.81 |
ENST00000547937.1 |
DCN |
decorin |
| chr12_-_15038779 | 2.10 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr3_+_69928256 | 1.14 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr12_-_92539614 | 1.07 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr10_+_70847852 | 1.05 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr2_+_233527443 | 0.95 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr4_-_138453606 | 0.92 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr20_+_45338126 | 0.89 |
ENST00000359271.2 |
SLC2A10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr1_+_163039143 | 0.85 |
ENST00000531057.1 ENST00000527809.1 ENST00000367908.4 |
RGS4 |
regulator of G-protein signaling 4 |
| chr1_+_221054584 | 0.75 |
ENST00000549319.1 |
HLX |
H2.0-like homeobox |
| chr3_+_148447887 | 0.73 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chrX_+_9431324 | 0.71 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr5_+_140529630 | 0.63 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chr1_-_24194771 | 0.61 |
ENST00000374479.3 |
FUCA1 |
fucosidase, alpha-L- 1, tissue |
| chr12_+_59989918 | 0.60 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr17_-_66951474 | 0.60 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr2_-_202563414 | 0.58 |
ENST00000409474.3 ENST00000315506.7 ENST00000359962.5 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr5_-_19988339 | 0.57 |
ENST00000382275.1 |
CDH18 |
cadherin 18, type 2 |
| chr6_-_117747015 | 0.57 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr2_+_88047606 | 0.56 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr12_-_15374343 | 0.56 |
ENST00000256953.2 ENST00000546331.1 |
RERG |
RAS-like, estrogen-regulated, growth inhibitor |
| chr6_+_122793058 | 0.54 |
ENST00000392491.2 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr2_+_109223595 | 0.53 |
ENST00000410093.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr14_-_74551096 | 0.51 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chrX_+_85969626 | 0.51 |
ENST00000484479.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr9_+_120466610 | 0.50 |
ENST00000394487.4 |
TLR4 |
toll-like receptor 4 |
| chr1_-_92371839 | 0.50 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr2_+_201450591 | 0.49 |
ENST00000374700.2 |
AOX1 |
aldehyde oxidase 1 |
| chr2_+_120770581 | 0.49 |
ENST00000263713.5 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr4_-_138453559 | 0.47 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr9_+_120466650 | 0.47 |
ENST00000355622.6 |
TLR4 |
toll-like receptor 4 |
| chr3_-_187455680 | 0.47 |
ENST00000438077.1 |
BCL6 |
B-cell CLL/lymphoma 6 |
| chr6_+_101846664 | 0.45 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr2_+_175260514 | 0.45 |
ENST00000424069.1 ENST00000427038.1 |
SCRN3 |
secernin 3 |
| chr9_-_21305312 | 0.44 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr14_+_88471468 | 0.41 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr18_-_25616519 | 0.41 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr1_+_150122034 | 0.40 |
ENST00000025469.6 ENST00000369124.4 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
| chr2_+_175260451 | 0.39 |
ENST00000458563.1 ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3 |
secernin 3 |
| chr1_+_185703513 | 0.39 |
ENST00000271588.4 ENST00000367492.2 |
HMCN1 |
hemicentin 1 |
| chr17_-_26662440 | 0.39 |
ENST00000578122.1 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr7_+_6617039 | 0.39 |
ENST00000405731.3 ENST00000396713.2 ENST00000396707.2 ENST00000335965.6 ENST00000396709.1 ENST00000483589.1 ENST00000396706.2 |
ZDHHC4 |
zinc finger, DHHC-type containing 4 |
| chr17_-_26662464 | 0.38 |
ENST00000579419.1 ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr14_-_74551172 | 0.38 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr21_-_31869451 | 0.38 |
ENST00000334058.2 |
KRTAP19-4 |
keratin associated protein 19-4 |
| chr17_-_19290483 | 0.37 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr7_+_149570049 | 0.37 |
ENST00000421974.2 ENST00000456496.2 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
| chr5_-_159846399 | 0.37 |
ENST00000297151.4 |
SLU7 |
SLU7 splicing factor homolog (S. cerevisiae) |
| chr8_+_70404996 | 0.35 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr19_-_58662139 | 0.35 |
ENST00000598312.1 |
ZNF329 |
zinc finger protein 329 |
| chr2_+_201170703 | 0.34 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr15_-_55657428 | 0.34 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr5_+_159436120 | 0.33 |
ENST00000522793.1 ENST00000231238.5 |
TTC1 |
tetratricopeptide repeat domain 1 |
| chr7_+_151038850 | 0.32 |
ENST00000355851.4 ENST00000566856.1 ENST00000470229.1 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
| chr6_+_21666633 | 0.31 |
ENST00000606851.1 |
CASC15 |
cancer susceptibility candidate 15 (non-protein coding) |
| chr1_+_178310581 | 0.30 |
ENST00000462775.1 |
RASAL2 |
RAS protein activator like 2 |
| chr2_+_65283506 | 0.30 |
ENST00000377990.2 |
CEP68 |
centrosomal protein 68kDa |
| chr10_-_25010795 | 0.30 |
ENST00000416305.1 ENST00000376410.2 |
ARHGAP21 |
Rho GTPase activating protein 21 |
| chr12_+_79439405 | 0.30 |
ENST00000552744.1 |
SYT1 |
synaptotagmin I |
| chr13_-_39564993 | 0.29 |
ENST00000423210.1 |
STOML3 |
stomatin (EPB72)-like 3 |
| chr6_+_108882069 | 0.29 |
ENST00000406360.1 |
FOXO3 |
forkhead box O3 |
| chr17_-_30185946 | 0.29 |
ENST00000579741.1 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
| chr7_+_106809406 | 0.29 |
ENST00000468410.1 ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1 |
HMG-box transcription factor 1 |
| chr5_-_146833222 | 0.28 |
ENST00000534907.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr1_+_78470530 | 0.27 |
ENST00000370763.5 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr2_+_109204743 | 0.27 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr5_-_131132614 | 0.27 |
ENST00000307968.7 ENST00000307954.8 |
FNIP1 |
folliculin interacting protein 1 |
| chr6_+_12717892 | 0.27 |
ENST00000379350.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr2_+_65283529 | 0.27 |
ENST00000546106.1 ENST00000537589.1 ENST00000260569.4 |
CEP68 |
centrosomal protein 68kDa |
| chr19_-_9546177 | 0.27 |
ENST00000592292.1 ENST00000588221.1 |
ZNF266 |
zinc finger protein 266 |
| chrX_-_106959631 | 0.26 |
ENST00000486554.1 ENST00000372390.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chrX_+_10124977 | 0.26 |
ENST00000380833.4 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
| chr13_-_49975632 | 0.26 |
ENST00000457041.1 ENST00000355854.4 |
CAB39L |
calcium binding protein 39-like |
| chr17_-_30185971 | 0.25 |
ENST00000378634.2 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
| chr2_+_12857043 | 0.25 |
ENST00000381465.2 |
TRIB2 |
tribbles pseudokinase 2 |
| chr2_+_109204909 | 0.25 |
ENST00000393310.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr2_+_210517895 | 0.24 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr2_-_87248975 | 0.23 |
ENST00000409310.2 ENST00000355705.3 |
PLGLB1 |
plasminogen-like B1 |
| chr10_-_14050522 | 0.23 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr2_-_224702740 | 0.23 |
ENST00000444408.1 |
AP1S3 |
adaptor-related protein complex 1, sigma 3 subunit |
| chr13_-_61989655 | 0.23 |
ENST00000409204.4 |
PCDH20 |
protocadherin 20 |
| chr19_+_18111927 | 0.22 |
ENST00000379656.3 |
ARRDC2 |
arrestin domain containing 2 |
| chr5_+_135468516 | 0.22 |
ENST00000507118.1 ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5 |
SMAD family member 5 |
| chr3_-_150920979 | 0.22 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr15_-_81616446 | 0.22 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr6_-_87804815 | 0.22 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr11_-_26593677 | 0.22 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr3_+_159570722 | 0.22 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chrX_+_120181457 | 0.22 |
ENST00000328078.1 |
GLUD2 |
glutamate dehydrogenase 2 |
| chr5_-_88119580 | 0.22 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr10_-_4285923 | 0.21 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chrX_+_54947229 | 0.21 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr12_-_92536433 | 0.21 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr2_+_120770645 | 0.20 |
ENST00000443902.2 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr12_+_21679220 | 0.20 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chr11_+_20044096 | 0.20 |
ENST00000533917.1 |
NAV2 |
neuron navigator 2 |
| chr1_+_74701062 | 0.20 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr4_+_74347400 | 0.20 |
ENST00000226355.3 |
AFM |
afamin |
| chr11_-_26593779 | 0.20 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr16_+_66442411 | 0.19 |
ENST00000499966.1 |
LINC00920 |
long intergenic non-protein coding RNA 920 |
| chr1_-_57431679 | 0.19 |
ENST00000371237.4 ENST00000535057.1 ENST00000543257.1 |
C8B |
complement component 8, beta polypeptide |
| chr5_+_140625147 | 0.19 |
ENST00000231173.3 |
PCDHB15 |
protocadherin beta 15 |
| chr1_-_20126365 | 0.19 |
ENST00000294543.6 ENST00000375122.2 |
TMCO4 |
transmembrane and coiled-coil domains 4 |
| chr2_-_175260368 | 0.18 |
ENST00000342016.3 ENST00000362053.5 |
CIR1 |
corepressor interacting with RBPJ, 1 |
| chr10_+_90672113 | 0.18 |
ENST00000371922.1 |
STAMBPL1 |
STAM binding protein-like 1 |
| chr5_+_140729649 | 0.18 |
ENST00000523390.1 |
PCDHGB1 |
protocadherin gamma subfamily B, 1 |
| chr16_-_80926457 | 0.18 |
ENST00000563626.1 ENST00000562231.1 |
RP11-314O13.1 |
RP11-314O13.1 |
| chr6_-_122792919 | 0.18 |
ENST00000339697.4 |
SERINC1 |
serine incorporator 1 |
| chr1_+_203765437 | 0.18 |
ENST00000550078.1 |
ZBED6 |
zinc finger, BED-type containing 6 |
| chr2_-_65593784 | 0.17 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr4_-_21950356 | 0.17 |
ENST00000447367.2 ENST00000382152.2 |
KCNIP4 |
Kv channel interacting protein 4 |
| chr5_+_141346385 | 0.17 |
ENST00000513019.1 ENST00000356143.1 |
RNF14 |
ring finger protein 14 |
| chr9_+_70856397 | 0.17 |
ENST00000360171.6 |
CBWD3 |
COBW domain containing 3 |
| chr2_+_166095898 | 0.17 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_+_88754113 | 0.17 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr12_-_7596735 | 0.17 |
ENST00000416109.2 ENST00000396630.1 ENST00000313599.3 |
CD163L1 |
CD163 molecule-like 1 |
| chr5_+_140792614 | 0.17 |
ENST00000398610.2 |
PCDHGA10 |
protocadherin gamma subfamily A, 10 |
| chr10_-_28623368 | 0.17 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_+_246729724 | 0.16 |
ENST00000366513.4 ENST00000366512.3 |
CNST |
consortin, connexin sorting protein |
| chr14_+_88851874 | 0.16 |
ENST00000393545.4 ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7 |
spermatogenesis associated 7 |
| chr2_-_32490859 | 0.16 |
ENST00000404025.2 |
NLRC4 |
NLR family, CARD domain containing 4 |
| chr6_+_161123270 | 0.16 |
ENST00000366924.2 ENST00000308192.9 ENST00000418964.1 |
PLG |
plasminogen |
| chr5_-_131132658 | 0.16 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr2_+_54342574 | 0.16 |
ENST00000303536.4 ENST00000394666.3 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr2_+_74120094 | 0.16 |
ENST00000409731.3 ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2 |
actin, gamma 2, smooth muscle, enteric |
| chr9_-_116163400 | 0.16 |
ENST00000277315.5 ENST00000448137.1 ENST00000409155.3 |
ALAD |
aminolevulinate dehydratase |
| chr20_-_30311703 | 0.16 |
ENST00000450273.1 ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1 |
BCL2-like 1 |
| chr2_+_61372226 | 0.16 |
ENST00000426997.1 |
C2orf74 |
chromosome 2 open reading frame 74 |
| chr20_+_56964169 | 0.15 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr3_-_58523010 | 0.15 |
ENST00000459701.2 ENST00000302819.5 |
ACOX2 |
acyl-CoA oxidase 2, branched chain |
| chr3_-_105588231 | 0.15 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr5_+_147774275 | 0.15 |
ENST00000513826.1 |
FBXO38 |
F-box protein 38 |
| chr3_-_127455200 | 0.15 |
ENST00000398101.3 |
MGLL |
monoglyceride lipase |
| chr1_+_174844645 | 0.15 |
ENST00000486220.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr10_-_49482907 | 0.15 |
ENST00000374201.3 ENST00000407470.4 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr5_-_153418407 | 0.15 |
ENST00000522858.1 ENST00000522634.1 ENST00000523705.1 ENST00000524246.1 ENST00000520313.1 ENST00000518102.1 ENST00000351797.4 ENST00000520667.1 ENST00000519808.1 ENST00000522395.1 |
FAM114A2 |
family with sequence similarity 114, member A2 |
| chr1_-_241803679 | 0.15 |
ENST00000331838.5 |
OPN3 |
opsin 3 |
| chr20_+_33104199 | 0.15 |
ENST00000357156.2 ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1 |
dynein, light chain, roadblock-type 1 |
| chr12_-_49582978 | 0.15 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chrX_+_153455547 | 0.14 |
ENST00000430054.1 |
OPN1MW |
opsin 1 (cone pigments), medium-wave-sensitive |
| chr15_-_40600026 | 0.14 |
ENST00000456256.2 ENST00000557821.1 |
PLCB2 |
phospholipase C, beta 2 |
| chr1_-_149982624 | 0.14 |
ENST00000417191.1 ENST00000369135.4 |
OTUD7B |
OTU domain containing 7B |
| chr1_-_241803649 | 0.14 |
ENST00000366554.2 |
OPN3 |
opsin 3 |
| chr22_-_43036607 | 0.14 |
ENST00000505920.1 |
ATP5L2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr7_-_6098770 | 0.14 |
ENST00000536084.1 ENST00000446699.1 ENST00000199389.6 |
EIF2AK1 |
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr4_+_88754069 | 0.14 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr9_-_70490107 | 0.14 |
ENST00000377395.4 ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5 |
COBW domain containing 5 |
| chr19_+_36630454 | 0.14 |
ENST00000246533.3 |
CAPNS1 |
calpain, small subunit 1 |
| chrX_-_47863348 | 0.14 |
ENST00000376943.3 ENST00000396965.1 ENST00000305127.6 |
ZNF182 |
zinc finger protein 182 |
| chr19_+_18208603 | 0.13 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr17_+_71229346 | 0.13 |
ENST00000535032.2 ENST00000582793.1 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr3_+_180319918 | 0.13 |
ENST00000296015.4 ENST00000491380.1 ENST00000412756.2 ENST00000382584.4 |
TTC14 |
tetratricopeptide repeat domain 14 |
| chrX_+_16668278 | 0.13 |
ENST00000380200.3 |
S100G |
S100 calcium binding protein G |
| chr3_-_151102529 | 0.13 |
ENST00000302632.3 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
| chr1_+_26036093 | 0.13 |
ENST00000374329.1 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
| chr11_-_26593649 | 0.13 |
ENST00000455601.2 |
MUC15 |
mucin 15, cell surface associated |
| chr4_-_76928641 | 0.13 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chr7_-_144435985 | 0.13 |
ENST00000549981.1 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr6_+_168418553 | 0.13 |
ENST00000354419.2 ENST00000351261.3 |
KIF25 |
kinesin family member 25 |
| chr4_-_140223614 | 0.12 |
ENST00000394223.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr14_+_105267250 | 0.12 |
ENST00000342537.7 |
ZBTB42 |
zinc finger and BTB domain containing 42 |
| chr13_-_103346854 | 0.12 |
ENST00000267273.6 |
METTL21C |
methyltransferase like 21C |
| chr19_+_57640011 | 0.12 |
ENST00000598197.1 |
USP29 |
ubiquitin specific peptidase 29 |
| chr11_-_9286921 | 0.12 |
ENST00000328194.3 |
DENND5A |
DENN/MADD domain containing 5A |
| chr19_-_52255107 | 0.12 |
ENST00000595042.1 ENST00000304748.4 |
FPR1 |
formyl peptide receptor 1 |
| chr9_-_86432547 | 0.12 |
ENST00000376365.3 ENST00000376371.2 |
GKAP1 |
G kinase anchoring protein 1 |
| chr17_-_39183452 | 0.12 |
ENST00000361883.5 |
KRTAP1-5 |
keratin associated protein 1-5 |
| chr8_+_26247878 | 0.12 |
ENST00000518611.1 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr3_+_119316689 | 0.12 |
ENST00000273371.4 |
PLA1A |
phospholipase A1 member A |
| chr12_-_76462713 | 0.12 |
ENST00000552056.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr3_-_169487617 | 0.12 |
ENST00000330368.2 |
ACTRT3 |
actin-related protein T3 |
| chr8_-_145754428 | 0.11 |
ENST00000527462.1 ENST00000313465.5 ENST00000524821.1 |
C8orf82 |
chromosome 8 open reading frame 82 |
| chr4_-_70826725 | 0.11 |
ENST00000353151.3 |
CSN2 |
casein beta |
| chr19_-_36523709 | 0.11 |
ENST00000592017.1 ENST00000360535.4 |
CLIP3 |
CAP-GLY domain containing linker protein 3 |
| chr11_-_47447767 | 0.11 |
ENST00000530651.1 ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr5_-_137674000 | 0.11 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr1_-_43855479 | 0.11 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr20_+_48552908 | 0.11 |
ENST00000244061.2 |
RNF114 |
ring finger protein 114 |
| chr16_+_28858004 | 0.11 |
ENST00000322610.8 |
SH2B1 |
SH2B adaptor protein 1 |
| chr8_+_100025476 | 0.11 |
ENST00000355155.1 ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B |
vacuolar protein sorting 13 homolog B (yeast) |
| chr19_-_40732594 | 0.11 |
ENST00000430325.2 ENST00000433940.1 |
CNTD2 |
cyclin N-terminal domain containing 2 |
| chr10_+_52152766 | 0.11 |
ENST00000596442.1 |
AC069547.2 |
Uncharacterized protein |
| chr11_-_47447970 | 0.11 |
ENST00000298852.3 ENST00000530912.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr2_-_3521518 | 0.11 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chr4_-_170679024 | 0.11 |
ENST00000393381.2 |
C4orf27 |
chromosome 4 open reading frame 27 |
| chr21_+_40817749 | 0.11 |
ENST00000380637.3 ENST00000380634.1 ENST00000458295.1 ENST00000440288.2 ENST00000380631.1 |
SH3BGR |
SH3 domain binding glutamic acid-rich protein |
| chr7_-_14028488 | 0.11 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr17_-_7531121 | 0.11 |
ENST00000573566.1 ENST00000269298.5 |
SAT2 |
spermidine/spermine N1-acetyltransferase family member 2 |
| chr1_+_84609944 | 0.11 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_+_119316721 | 0.10 |
ENST00000488919.1 ENST00000495992.1 |
PLA1A |
phospholipase A1 member A |
| chr1_+_196621002 | 0.10 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr20_+_48429233 | 0.10 |
ENST00000417961.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr12_-_76879852 | 0.10 |
ENST00000548341.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr6_-_161085291 | 0.10 |
ENST00000316300.5 |
LPA |
lipoprotein, Lp(a) |
| chr9_-_13175823 | 0.10 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr1_+_161123536 | 0.10 |
ENST00000368003.5 |
UFC1 |
ubiquitin-fold modifier conjugating enzyme 1 |
| chr1_-_169555779 | 0.10 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr8_-_100025238 | 0.10 |
ENST00000521696.1 |
RP11-410L14.2 |
RP11-410L14.2 |
| chr1_-_16763685 | 0.10 |
ENST00000540400.1 |
SPATA21 |
spermatogenesis associated 21 |
| chr7_-_38389573 | 0.10 |
ENST00000390344.2 |
TRGV5 |
T cell receptor gamma variable 5 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.3 | 1.0 | GO:0070428 | negative regulation of interleukin-23 production(GO:0032707) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.2 | 0.9 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.2 | 2.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.7 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 0.7 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.2 | 0.5 | GO:0046113 | purine nucleobase catabolic process(GO:0006145) xanthine metabolic process(GO:0046110) nucleobase catabolic process(GO:0046113) |
| 0.2 | 0.5 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.5 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.4 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 2.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 1.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.6 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.6 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.2 | GO:0070541 | response to platinum ion(GO:0070541) cellular response to lead ion(GO:0071284) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.3 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.9 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.4 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.9 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 1.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:1903509 | glycolipid metabolic process(GO:0006664) liposaccharide metabolic process(GO:1903509) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.2 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.0 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 2.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0035701 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.4 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 2.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.5 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 0.6 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 0.8 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.6 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 3.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 2.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |