Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for FOXD3_FOXI1_FOXF1
Z-value: 1.09
Transcription factors associated with FOXD3_FOXI1_FOXF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
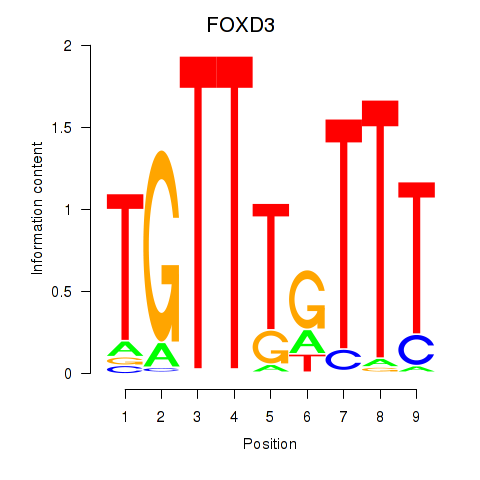
FOXD3
|
ENSG00000187140.4 | FOXD3 |
|
FOXI1
|
ENSG00000168269.7 | FOXI1 |
|
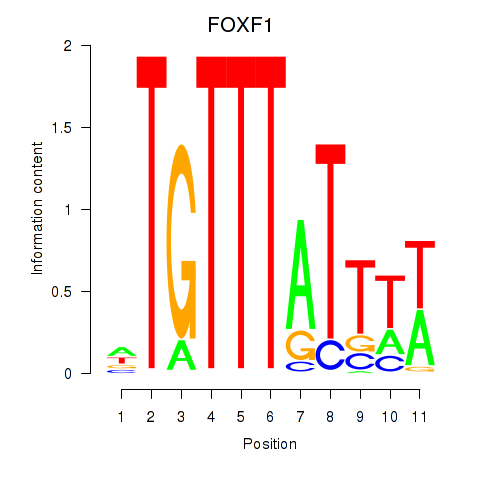
FOXF1
|
ENSG00000103241.5 | FOXF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXI1 | hg19_v2_chr5_+_169532896_169532917 | 0.86 | 6.3e-03 | Click! |
| FOXF1 | hg19_v2_chr16_+_86544113_86544145 | -0.67 | 7.2e-02 | Click! |
| FOXD3 | hg19_v2_chr1_+_63788730_63788730 | -0.35 | 3.9e-01 | Click! |
Activity profile of FOXD3_FOXI1_FOXF1 motif
Sorted Z-values of FOXD3_FOXI1_FOXF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXD3_FOXI1_FOXF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_97597148 | 2.27 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr8_-_108510224 | 1.87 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr12_-_15038779 | 1.70 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr17_-_19290483 | 1.50 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr1_-_57045228 | 1.32 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr18_-_52626622 | 1.21 |
ENST00000591504.1 |
CCDC68 |
coiled-coil domain containing 68 |
| chr14_-_92414055 | 1.17 |
ENST00000342058.4 |
FBLN5 |
fibulin 5 |
| chr4_-_186696425 | 1.11 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr9_-_95244781 | 0.96 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr8_+_104384616 | 0.93 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr3_-_114343039 | 0.87 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr11_-_111781610 | 0.86 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr5_-_111091948 | 0.80 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr9_-_89562104 | 0.73 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chr17_+_57408994 | 0.72 |
ENST00000312655.4 |
YPEL2 |
yippee-like 2 (Drosophila) |
| chr11_-_111781554 | 0.70 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chrX_+_22050546 | 0.69 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr11_-_111781454 | 0.69 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr5_+_32788945 | 0.68 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr5_-_172198190 | 0.68 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr3_+_45067659 | 0.66 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chrX_+_66764375 | 0.66 |
ENST00000374690.3 |
AR |
androgen receptor |
| chr3_-_195310802 | 0.65 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr12_-_92539614 | 0.64 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr8_-_120685608 | 0.63 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr8_+_104383728 | 0.63 |
ENST00000330295.5 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr2_+_166095898 | 0.60 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr6_+_101846664 | 0.60 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr12_-_91576561 | 0.60 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr4_+_124317940 | 0.60 |
ENST00000505319.1 ENST00000339241.1 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr8_+_97506033 | 0.58 |
ENST00000518385.1 |
SDC2 |
syndecan 2 |
| chr2_-_72374948 | 0.57 |
ENST00000546307.1 ENST00000474509.1 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr1_+_159141397 | 0.57 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr2_+_128175997 | 0.54 |
ENST00000234071.3 ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr5_-_19988339 | 0.54 |
ENST00000382275.1 |
CDH18 |
cadherin 18, type 2 |
| chr18_-_56296182 | 0.52 |
ENST00000361673.3 |
ALPK2 |
alpha-kinase 2 |
| chrX_+_16964985 | 0.51 |
ENST00000303843.7 |
REPS2 |
RALBP1 associated Eps domain containing 2 |
| chr3_-_114035026 | 0.49 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr8_-_120651020 | 0.49 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr1_-_72566613 | 0.48 |
ENST00000306821.3 |
NEGR1 |
neuronal growth regulator 1 |
| chr8_+_75736761 | 0.48 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr1_+_197886461 | 0.47 |
ENST00000367388.3 ENST00000337020.2 ENST00000367387.4 |
LHX9 |
LIM homeobox 9 |
| chr2_-_175711133 | 0.46 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr3_-_186080012 | 0.45 |
ENST00000544847.1 ENST00000265022.3 |
DGKG |
diacylglycerol kinase, gamma 90kDa |
| chr11_+_63304273 | 0.44 |
ENST00000439013.2 ENST00000255688.3 |
RARRES3 |
retinoic acid receptor responder (tazarotene induced) 3 |
| chr12_-_91574142 | 0.43 |
ENST00000547937.1 |
DCN |
decorin |
| chr1_+_223101757 | 0.43 |
ENST00000284476.6 |
DISP1 |
dispatched homolog 1 (Drosophila) |
| chr5_-_20575959 | 0.42 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr5_+_119799927 | 0.42 |
ENST00000407149.2 ENST00000379551.2 |
PRR16 |
proline rich 16 |
| chr3_+_99357319 | 0.41 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr8_+_79428539 | 0.41 |
ENST00000352966.5 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr6_+_136172820 | 0.41 |
ENST00000308191.6 |
PDE7B |
phosphodiesterase 7B |
| chr1_+_164528866 | 0.41 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr2_+_33359646 | 0.40 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 0.40 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_152214098 | 0.40 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr9_-_95298314 | 0.40 |
ENST00000344604.5 ENST00000375540.1 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
| chr17_-_66951474 | 0.39 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr3_-_16524357 | 0.39 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr2_-_152146385 | 0.38 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr3_+_69812877 | 0.38 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr6_-_154751629 | 0.37 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr8_-_93107443 | 0.37 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr4_-_175443943 | 0.37 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr1_+_164529004 | 0.37 |
ENST00000559240.1 ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr7_+_134430212 | 0.37 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr5_-_158526756 | 0.36 |
ENST00000313708.6 ENST00000517373.1 |
EBF1 |
early B-cell factor 1 |
| chr12_-_91576429 | 0.36 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr21_+_17792672 | 0.36 |
ENST00000602620.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr8_+_52730143 | 0.35 |
ENST00000415643.1 |
AC090186.1 |
Uncharacterized protein |
| chr12_+_79439405 | 0.35 |
ENST00000552744.1 |
SYT1 |
synaptotagmin I |
| chr12_-_71551652 | 0.34 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr11_+_64085560 | 0.34 |
ENST00000265462.4 ENST00000352435.4 ENST00000347941.4 |
PRDX5 |
peroxiredoxin 5 |
| chr2_-_2334888 | 0.34 |
ENST00000428368.2 ENST00000399161.2 |
MYT1L |
myelin transcription factor 1-like |
| chr12_-_56106060 | 0.34 |
ENST00000452168.2 |
ITGA7 |
integrin, alpha 7 |
| chr1_-_227505289 | 0.33 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr10_+_31610064 | 0.33 |
ENST00000446923.2 ENST00000559476.1 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr3_+_114012819 | 0.33 |
ENST00000383671.3 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
| chr12_-_15374343 | 0.32 |
ENST00000256953.2 ENST00000546331.1 |
RERG |
RAS-like, estrogen-regulated, growth inhibitor |
| chr18_-_25616519 | 0.32 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr6_-_25830785 | 0.32 |
ENST00000468082.1 |
SLC17A1 |
solute carrier family 17 (organic anion transporter), member 1 |
| chr13_-_67802549 | 0.31 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr5_+_156712372 | 0.31 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr11_-_102826434 | 0.31 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr12_+_10365404 | 0.31 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr2_+_233497931 | 0.30 |
ENST00000264059.3 |
EFHD1 |
EF-hand domain family, member D1 |
| chr8_-_60031762 | 0.30 |
ENST00000361421.1 |
TOX |
thymocyte selection-associated high mobility group box |
| chr5_-_127674883 | 0.30 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr1_+_150122034 | 0.30 |
ENST00000025469.6 ENST00000369124.4 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
| chr1_-_19811132 | 0.30 |
ENST00000433834.1 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
| chr5_-_158526693 | 0.30 |
ENST00000380654.4 |
EBF1 |
early B-cell factor 1 |
| chr6_-_152489484 | 0.30 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr5_-_111093167 | 0.30 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr5_+_40841276 | 0.30 |
ENST00000254691.5 |
CARD6 |
caspase recruitment domain family, member 6 |
| chr2_+_166150541 | 0.29 |
ENST00000283256.6 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr6_+_89790490 | 0.29 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr2_-_145275228 | 0.29 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chrX_+_100743031 | 0.29 |
ENST00000423738.3 |
ARMCX4 |
armadillo repeat containing, X-linked 4 |
| chr4_+_74347400 | 0.28 |
ENST00000226355.3 |
AFM |
afamin |
| chr6_-_152639479 | 0.28 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_1297157 | 0.28 |
ENST00000477278.2 |
MXRA8 |
matrix-remodelling associated 8 |
| chr21_+_17791838 | 0.28 |
ENST00000453910.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr1_+_104159999 | 0.28 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr12_+_25348186 | 0.28 |
ENST00000555711.1 ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5 |
LYR motif containing 5 |
| chr4_-_186732048 | 0.28 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr2_-_188419200 | 0.28 |
ENST00000233156.3 ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr21_+_17791648 | 0.28 |
ENST00000602892.1 ENST00000418813.2 ENST00000435697.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr15_-_93632421 | 0.27 |
ENST00000329082.7 |
RGMA |
repulsive guidance molecule family member a |
| chr1_-_92371839 | 0.27 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr8_-_93107696 | 0.27 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_+_157154578 | 0.27 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr1_+_207277590 | 0.27 |
ENST00000367070.3 |
C4BPA |
complement component 4 binding protein, alpha |
| chr4_+_129732467 | 0.27 |
ENST00000413543.2 |
PHF17 |
jade family PHD finger 1 |
| chr17_+_79953310 | 0.27 |
ENST00000582355.2 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr7_+_151038850 | 0.27 |
ENST00000355851.4 ENST00000566856.1 ENST00000470229.1 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
| chrX_-_102531717 | 0.26 |
ENST00000372680.1 |
TCEAL5 |
transcription elongation factor A (SII)-like 5 |
| chrX_+_22056165 | 0.26 |
ENST00000535894.1 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr6_-_88875654 | 0.26 |
ENST00000535130.1 |
CNR1 |
cannabinoid receptor 1 (brain) |
| chr21_+_30502806 | 0.26 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr1_+_171060018 | 0.26 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr12_+_25348139 | 0.26 |
ENST00000557540.2 ENST00000381356.4 |
LYRM5 |
LYR motif containing 5 |
| chr4_-_175443484 | 0.26 |
ENST00000514584.1 ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr8_-_93107827 | 0.26 |
ENST00000520724.1 ENST00000518844.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_-_153599732 | 0.25 |
ENST00000392623.1 |
S100A13 |
S100 calcium binding protein A13 |
| chr11_+_5710919 | 0.25 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr7_+_48211048 | 0.25 |
ENST00000435803.1 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr10_-_31288398 | 0.25 |
ENST00000538351.2 |
ZNF438 |
zinc finger protein 438 |
| chr12_+_51318513 | 0.25 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr2_+_158114051 | 0.25 |
ENST00000259056.4 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr5_+_161495038 | 0.24 |
ENST00000393933.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr2_-_188419078 | 0.24 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_-_106054952 | 0.24 |
ENST00000336660.5 ENST00000393352.3 ENST00000607522.1 |
FHL2 |
four and a half LIM domains 2 |
| chr7_+_18535854 | 0.24 |
ENST00000401921.1 |
HDAC9 |
histone deacetylase 9 |
| chr8_+_70404996 | 0.24 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr3_-_114477787 | 0.23 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr10_+_111967345 | 0.23 |
ENST00000332674.5 ENST00000453116.1 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr3_-_121379739 | 0.23 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr6_-_131291572 | 0.23 |
ENST00000529208.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr2_+_201994569 | 0.23 |
ENST00000457277.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr16_-_67260691 | 0.23 |
ENST00000447579.1 ENST00000393992.1 ENST00000424285.1 |
LRRC29 |
leucine rich repeat containing 29 |
| chr5_+_139027877 | 0.23 |
ENST00000302517.3 |
CXXC5 |
CXXC finger protein 5 |
| chr4_+_86396265 | 0.23 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr15_-_55657428 | 0.23 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr6_-_133084580 | 0.23 |
ENST00000525270.1 ENST00000530536.1 ENST00000524919.1 |
VNN2 |
vanin 2 |
| chr3_-_165555200 | 0.23 |
ENST00000479451.1 ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE |
butyrylcholinesterase |
| chr9_-_20622478 | 0.23 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr7_-_92777606 | 0.22 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr1_+_162602244 | 0.22 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chrX_+_85969626 | 0.22 |
ENST00000484479.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr4_-_159094194 | 0.22 |
ENST00000592057.1 ENST00000585682.1 ENST00000393807.5 |
FAM198B |
family with sequence similarity 198, member B |
| chr3_-_187454281 | 0.22 |
ENST00000232014.4 |
BCL6 |
B-cell CLL/lymphoma 6 |
| chr7_-_27142290 | 0.22 |
ENST00000222718.5 |
HOXA2 |
homeobox A2 |
| chr7_+_7811992 | 0.22 |
ENST00000406829.1 |
RPA3-AS1 |
RPA3 antisense RNA 1 |
| chr6_+_89790459 | 0.22 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr4_+_38869298 | 0.21 |
ENST00000510213.1 ENST00000515037.1 |
FAM114A1 |
family with sequence similarity 114, member A1 |
| chr3_+_141105705 | 0.21 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr4_-_175443788 | 0.21 |
ENST00000541923.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chrX_-_154493791 | 0.21 |
ENST00000369454.3 |
RAB39B |
RAB39B, member RAS oncogene family |
| chr11_-_62323702 | 0.21 |
ENST00000530285.1 |
AHNAK |
AHNAK nucleoprotein |
| chr6_-_167276033 | 0.21 |
ENST00000503859.1 ENST00000506565.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr8_-_72274467 | 0.21 |
ENST00000340726.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr7_-_14029283 | 0.21 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr2_+_109223595 | 0.21 |
ENST00000410093.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr17_+_17206635 | 0.21 |
ENST00000389022.4 |
NT5M |
5',3'-nucleotidase, mitochondrial |
| chr3_+_69812701 | 0.21 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr3_+_154797428 | 0.20 |
ENST00000460393.1 |
MME |
membrane metallo-endopeptidase |
| chr1_+_59775752 | 0.20 |
ENST00000371212.1 |
FGGY |
FGGY carbohydrate kinase domain containing |
| chr6_-_117747015 | 0.20 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr5_+_140571902 | 0.20 |
ENST00000239446.4 |
PCDHB10 |
protocadherin beta 10 |
| chr6_+_72926145 | 0.20 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr21_-_28338732 | 0.20 |
ENST00000284987.5 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chr5_+_145718587 | 0.20 |
ENST00000230732.4 |
POU4F3 |
POU class 4 homeobox 3 |
| chr10_+_95848824 | 0.20 |
ENST00000371385.3 ENST00000371375.1 |
PLCE1 |
phospholipase C, epsilon 1 |
| chrX_-_101410762 | 0.20 |
ENST00000543160.1 ENST00000333643.3 |
BEX5 |
brain expressed, X-linked 5 |
| chr14_-_61190754 | 0.20 |
ENST00000216513.4 |
SIX4 |
SIX homeobox 4 |
| chr16_+_4421841 | 0.20 |
ENST00000304735.3 |
VASN |
vasorin |
| chrX_+_55744166 | 0.20 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr4_-_84035905 | 0.20 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chr7_+_134551583 | 0.20 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr8_-_72274095 | 0.20 |
ENST00000303824.7 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr10_-_90712520 | 0.20 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr6_+_114178512 | 0.20 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr7_+_142985308 | 0.19 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr18_+_3449330 | 0.19 |
ENST00000549253.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chrX_+_55744228 | 0.19 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr17_-_67138015 | 0.19 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr5_-_140700322 | 0.19 |
ENST00000313368.5 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr15_-_52944231 | 0.19 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr19_-_39322299 | 0.19 |
ENST00000601094.1 ENST00000595567.1 ENST00000602115.1 ENST00000601778.1 ENST00000597205.1 ENST00000595470.1 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
| chr12_-_10605929 | 0.19 |
ENST00000347831.5 ENST00000359151.3 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr12_-_22487618 | 0.19 |
ENST00000404299.3 ENST00000396037.4 |
ST8SIA1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr11_-_102401469 | 0.19 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr8_-_12668962 | 0.19 |
ENST00000534827.1 |
RP11-252C15.1 |
RP11-252C15.1 |
| chr12_+_59989791 | 0.18 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr5_-_133510456 | 0.18 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr8_-_13372253 | 0.18 |
ENST00000316609.5 |
DLC1 |
deleted in liver cancer 1 |
| chr6_-_82462425 | 0.18 |
ENST00000369754.3 ENST00000320172.6 ENST00000369756.3 |
FAM46A |
family with sequence similarity 46, member A |
| chr2_-_211168332 | 0.18 |
ENST00000341685.4 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr12_+_99038919 | 0.18 |
ENST00000551964.1 |
APAF1 |
apoptotic peptidase activating factor 1 |
| chr3_+_152017924 | 0.18 |
ENST00000465907.2 ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.0 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 0.7 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 1.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.6 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 0.7 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.2 | 0.6 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 2.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.9 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 0.5 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.3 | GO:0060939 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.1 | 2.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.5 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 1.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.8 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 1.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.4 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.2 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.2 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.7 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 1.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.2 | GO:1902725 | skeletal muscle fiber differentiation(GO:0098528) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.3 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.2 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.1 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.1 | GO:0071335 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.1 | 0.6 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.5 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.4 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.8 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.5 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.3 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.4 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.0 | 0.3 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.3 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.3 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.3 | GO:0046324 | regulation of glucose transport(GO:0010827) regulation of glucose import(GO:0046324) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 1.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.0 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.3 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 0.1 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.2 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0060318 | regulation of definitive erythrocyte differentiation(GO:0010724) definitive erythrocyte differentiation(GO:0060318) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.3 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.1 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) Schwann cell proliferation(GO:0014010) negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 | 0.2 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.1 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.0 | GO:0050968 | thermoception(GO:0050955) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.0 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.0 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.0 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.0 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 2.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.6 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.4 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.5 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 3.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 1.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.8 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.2 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 2.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.3 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.3 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 1.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.3 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 1.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0001026 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 3.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 3.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |