Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
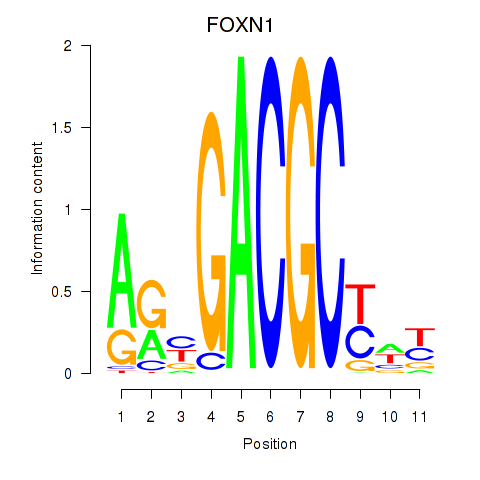
Results for FOXN1
Z-value: 0.46
Transcription factors associated with FOXN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXN1
|
ENSG00000109101.3 | FOXN1 |
Activity profile of FOXN1 motif
Sorted Z-values of FOXN1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXN1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_7942335 | 0.43 |
ENST00000380183.4 ENST00000572022.1 ENST00000380173.2 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr12_-_12715266 | 0.42 |
ENST00000228862.2 |
DUSP16 |
dual specificity phosphatase 16 |
| chr17_+_7942424 | 0.42 |
ENST00000573359.1 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr18_+_3448455 | 0.38 |
ENST00000549780.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr14_+_61788429 | 0.35 |
ENST00000332981.5 |
PRKCH |
protein kinase C, eta |
| chr3_-_38691119 | 0.27 |
ENST00000333535.4 ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A |
sodium channel, voltage-gated, type V, alpha subunit |
| chr10_+_104180580 | 0.26 |
ENST00000425536.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chrX_+_152953505 | 0.23 |
ENST00000253122.5 |
SLC6A8 |
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr17_+_44928946 | 0.23 |
ENST00000290015.2 ENST00000393461.2 |
WNT9B |
wingless-type MMTV integration site family, member 9B |
| chr6_+_27215471 | 0.22 |
ENST00000421826.2 |
PRSS16 |
protease, serine, 16 (thymus) |
| chr8_+_86089460 | 0.22 |
ENST00000418930.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr16_-_57831914 | 0.21 |
ENST00000421376.2 |
KIFC3 |
kinesin family member C3 |
| chr6_+_27215494 | 0.21 |
ENST00000230582.3 |
PRSS16 |
protease, serine, 16 (thymus) |
| chr14_-_92302784 | 0.21 |
ENST00000340892.5 ENST00000360594.5 |
TC2N |
tandem C2 domains, nuclear |
| chr13_-_95364389 | 0.21 |
ENST00000376945.2 |
SOX21 |
SRY (sex determining region Y)-box 21 |
| chr20_+_61273797 | 0.21 |
ENST00000217159.1 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
| chrX_-_117107542 | 0.21 |
ENST00000371878.1 |
KLHL13 |
kelch-like family member 13 |
| chr12_-_102224457 | 0.20 |
ENST00000549165.1 ENST00000549940.1 ENST00000392919.4 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chrX_+_68048803 | 0.20 |
ENST00000204961.4 |
EFNB1 |
ephrin-B1 |
| chrX_-_117107680 | 0.20 |
ENST00000447671.2 ENST00000262820.3 |
KLHL13 |
kelch-like family member 13 |
| chr4_+_81256871 | 0.18 |
ENST00000358105.3 ENST00000508675.1 |
C4orf22 |
chromosome 4 open reading frame 22 |
| chrX_-_83442915 | 0.18 |
ENST00000262752.2 ENST00000543399.1 |
RPS6KA6 |
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chrX_+_153237740 | 0.18 |
ENST00000369982.4 |
TMEM187 |
transmembrane protein 187 |
| chr7_-_130353553 | 0.17 |
ENST00000330992.7 ENST00000445977.2 |
COPG2 |
coatomer protein complex, subunit gamma 2 |
| chr15_+_45422178 | 0.17 |
ENST00000389037.3 ENST00000558322.1 |
DUOX1 |
dual oxidase 1 |
| chr15_-_59665062 | 0.16 |
ENST00000288235.4 |
MYO1E |
myosin IE |
| chr17_+_6916957 | 0.16 |
ENST00000547302.2 |
RNASEK-C17orf49 |
RNASEK-C17orf49 readthrough |
| chr15_+_45422131 | 0.16 |
ENST00000321429.4 |
DUOX1 |
dual oxidase 1 |
| chr14_-_92302825 | 0.16 |
ENST00000556018.1 |
TC2N |
tandem C2 domains, nuclear |
| chr3_-_150481164 | 0.16 |
ENST00000312960.3 |
SIAH2 |
siah E3 ubiquitin protein ligase 2 |
| chr12_+_51632638 | 0.16 |
ENST00000549732.2 |
DAZAP2 |
DAZ associated protein 2 |
| chr19_+_48867652 | 0.15 |
ENST00000344846.2 |
SYNGR4 |
synaptogyrin 4 |
| chr9_-_99775862 | 0.15 |
ENST00000602917.1 ENST00000375223.4 |
HIATL2 |
hippocampus abundant transcript-like 2 |
| chr2_-_28113217 | 0.15 |
ENST00000444339.2 |
RBKS |
ribokinase |
| chr1_+_82266053 | 0.15 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr8_+_38243967 | 0.15 |
ENST00000524874.1 ENST00000379957.4 ENST00000523983.2 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr15_+_92937144 | 0.14 |
ENST00000539113.1 ENST00000555434.1 |
ST8SIA2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr10_+_127585093 | 0.14 |
ENST00000368695.1 ENST00000368693.1 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
| chr4_-_10023095 | 0.14 |
ENST00000264784.3 |
SLC2A9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chrX_+_101380642 | 0.13 |
ENST00000372780.1 ENST00000329035.2 |
TCEAL2 |
transcription elongation factor A (SII)-like 2 |
| chr11_-_108464321 | 0.13 |
ENST00000265843.4 |
EXPH5 |
exophilin 5 |
| chrX_-_48815633 | 0.13 |
ENST00000428668.2 |
OTUD5 |
OTU domain containing 5 |
| chr19_+_11039391 | 0.13 |
ENST00000270502.6 |
C19orf52 |
chromosome 19 open reading frame 52 |
| chr12_+_70760056 | 0.13 |
ENST00000258111.4 |
KCNMB4 |
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr8_+_38243821 | 0.12 |
ENST00000519476.2 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr12_+_51632600 | 0.12 |
ENST00000549555.1 ENST00000439799.2 ENST00000425012.2 |
DAZAP2 |
DAZ associated protein 2 |
| chr1_-_25256368 | 0.12 |
ENST00000308873.6 |
RUNX3 |
runt-related transcription factor 3 |
| chr1_-_78444776 | 0.12 |
ENST00000370767.1 ENST00000421641.1 |
FUBP1 |
far upstream element (FUSE) binding protein 1 |
| chr17_+_40913264 | 0.12 |
ENST00000587142.1 ENST00000588576.1 |
RAMP2 |
receptor (G protein-coupled) activity modifying protein 2 |
| chr20_+_825275 | 0.12 |
ENST00000541082.1 |
FAM110A |
family with sequence similarity 110, member A |
| chr11_+_64059464 | 0.11 |
ENST00000394525.2 |
KCNK4 |
potassium channel, subfamily K, member 4 |
| chr3_-_52443799 | 0.11 |
ENST00000470173.1 ENST00000296288.5 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr1_-_78444738 | 0.11 |
ENST00000436586.2 ENST00000370768.2 |
FUBP1 |
far upstream element (FUSE) binding protein 1 |
| chr7_+_96747030 | 0.11 |
ENST00000360382.4 |
ACN9 |
ACN9 homolog (S. cerevisiae) |
| chr2_-_165697920 | 0.11 |
ENST00000342193.4 ENST00000375458.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr4_+_119200215 | 0.11 |
ENST00000602573.1 |
SNHG8 |
small nucleolar RNA host gene 8 (non-protein coding) |
| chr8_-_30515693 | 0.11 |
ENST00000355904.4 |
GTF2E2 |
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr14_+_96968802 | 0.11 |
ENST00000556619.1 ENST00000392990.2 |
PAPOLA |
poly(A) polymerase alpha |
| chr5_+_112312399 | 0.10 |
ENST00000515408.1 ENST00000513585.1 |
DCP2 |
decapping mRNA 2 |
| chr17_+_36508111 | 0.10 |
ENST00000331159.5 ENST00000577233.1 |
SOCS7 |
suppressor of cytokine signaling 7 |
| chr18_+_48405419 | 0.10 |
ENST00000321341.5 |
ME2 |
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr19_+_33463210 | 0.10 |
ENST00000590281.1 |
C19orf40 |
chromosome 19 open reading frame 40 |
| chr22_+_20104947 | 0.10 |
ENST00000402752.1 |
RANBP1 |
RAN binding protein 1 |
| chr14_+_96968707 | 0.10 |
ENST00000216277.8 ENST00000557320.1 ENST00000557471.1 |
PAPOLA |
poly(A) polymerase alpha |
| chr14_+_37126765 | 0.10 |
ENST00000402703.2 |
PAX9 |
paired box 9 |
| chr1_-_46152174 | 0.10 |
ENST00000290795.3 ENST00000355105.3 |
GPBP1L1 |
GC-rich promoter binding protein 1-like 1 |
| chr5_+_154238149 | 0.10 |
ENST00000519430.1 ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr8_+_38243951 | 0.10 |
ENST00000297720.5 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr19_-_11639910 | 0.10 |
ENST00000588998.1 ENST00000586149.1 |
ECSIT |
ECSIT signalling integrator |
| chr5_-_131892501 | 0.09 |
ENST00000450655.1 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr12_+_50451331 | 0.09 |
ENST00000228468.4 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chr5_+_176873789 | 0.09 |
ENST00000323249.3 ENST00000502922.1 |
PRR7 |
proline rich 7 (synaptic) |
| chr10_-_27529486 | 0.09 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr16_-_30022293 | 0.09 |
ENST00000565273.1 ENST00000567332.2 ENST00000350119.4 |
DOC2A |
double C2-like domains, alpha |
| chr6_+_7590413 | 0.09 |
ENST00000342415.5 |
SNRNP48 |
small nuclear ribonucleoprotein 48kDa (U11/U12) |
| chr16_-_30538079 | 0.09 |
ENST00000562803.1 |
ZNF768 |
zinc finger protein 768 |
| chr3_+_52280173 | 0.09 |
ENST00000296487.4 |
PPM1M |
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr15_-_52472078 | 0.09 |
ENST00000396335.4 ENST00000560116.1 ENST00000358784.7 |
GNB5 |
guanine nucleotide binding protein (G protein), beta 5 |
| chr1_+_153606513 | 0.09 |
ENST00000368694.3 |
CHTOP |
chromatin target of PRMT1 |
| chr19_-_47220335 | 0.09 |
ENST00000601806.1 ENST00000593363.1 ENST00000598633.1 ENST00000595515.1 ENST00000433867.1 |
PRKD2 |
protein kinase D2 |
| chr12_+_51632666 | 0.09 |
ENST00000604900.1 |
DAZAP2 |
DAZ associated protein 2 |
| chr19_-_11639931 | 0.09 |
ENST00000592312.1 ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT |
ECSIT signalling integrator |
| chr2_+_234160217 | 0.09 |
ENST00000392017.4 ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
| chr20_-_58515344 | 0.09 |
ENST00000370996.3 |
PPP1R3D |
protein phosphatase 1, regulatory subunit 3D |
| chr16_+_1203194 | 0.09 |
ENST00000348261.5 ENST00000358590.4 |
CACNA1H |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr2_+_241508039 | 0.09 |
ENST00000270357.4 |
RNPEPL1 |
arginyl aminopeptidase (aminopeptidase B)-like 1 |
| chr5_+_122847908 | 0.09 |
ENST00000511130.2 ENST00000512718.3 |
CSNK1G3 |
casein kinase 1, gamma 3 |
| chr22_-_42084863 | 0.08 |
ENST00000401959.1 ENST00000355257.3 |
NHP2L1 |
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr3_+_52280220 | 0.08 |
ENST00000409502.3 ENST00000323588.4 |
PPM1M |
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr15_-_78423863 | 0.08 |
ENST00000539011.1 |
CIB2 |
calcium and integrin binding family member 2 |
| chr16_-_30022735 | 0.08 |
ENST00000564944.1 |
DOC2A |
double C2-like domains, alpha |
| chr19_-_42806919 | 0.08 |
ENST00000595530.1 ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr2_-_152684977 | 0.08 |
ENST00000428992.2 ENST00000295087.8 |
ARL5A |
ADP-ribosylation factor-like 5A |
| chr2_+_234160340 | 0.08 |
ENST00000417017.1 ENST00000392020.4 ENST00000392018.1 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
| chr3_-_120461353 | 0.08 |
ENST00000483733.1 |
RABL3 |
RAB, member of RAS oncogene family-like 3 |
| chr2_-_31360887 | 0.08 |
ENST00000420311.2 ENST00000356174.3 ENST00000324589.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr9_+_102668915 | 0.08 |
ENST00000259400.6 ENST00000531035.1 ENST00000525640.1 ENST00000534052.1 ENST00000526607.1 |
STX17 |
syntaxin 17 |
| chr18_+_9708162 | 0.08 |
ENST00000578921.1 |
RAB31 |
RAB31, member RAS oncogene family |
| chr9_-_99382065 | 0.08 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chr1_-_59012365 | 0.08 |
ENST00000456980.1 ENST00000482274.2 ENST00000453710.1 ENST00000419242.1 ENST00000358603.2 ENST00000371226.3 ENST00000426139.1 |
OMA1 |
OMA1 zinc metallopeptidase |
| chr12_+_56110247 | 0.08 |
ENST00000551926.1 |
BLOC1S1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr4_+_119199904 | 0.08 |
ENST00000602483.1 ENST00000602819.1 |
SNHG8 |
small nucleolar RNA host gene 8 (non-protein coding) |
| chr18_+_48405463 | 0.08 |
ENST00000382927.3 |
ME2 |
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr12_+_3600356 | 0.08 |
ENST00000382622.3 |
PRMT8 |
protein arginine methyltransferase 8 |
| chr15_+_72766651 | 0.08 |
ENST00000379887.4 |
ARIH1 |
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr4_+_164415785 | 0.08 |
ENST00000513272.1 ENST00000513134.1 |
TMA16 |
translation machinery associated 16 homolog (S. cerevisiae) |
| chr11_+_66360665 | 0.08 |
ENST00000310190.4 |
CCS |
copper chaperone for superoxide dismutase |
| chr1_-_28241024 | 0.07 |
ENST00000313433.7 ENST00000444045.1 |
RPA2 |
replication protein A2, 32kDa |
| chr11_-_9025541 | 0.07 |
ENST00000525100.1 ENST00000309166.3 ENST00000531090.1 |
NRIP3 |
nuclear receptor interacting protein 3 |
| chr3_-_48936272 | 0.07 |
ENST00000544097.1 ENST00000430379.1 ENST00000319017.4 |
SLC25A20 |
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr5_+_32585605 | 0.07 |
ENST00000265073.4 ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
| chr15_+_99791567 | 0.07 |
ENST00000558879.1 ENST00000301981.3 ENST00000422500.2 ENST00000447360.2 ENST00000442993.2 |
LRRC28 |
leucine rich repeat containing 28 |
| chr1_+_206138457 | 0.07 |
ENST00000367128.3 ENST00000431655.2 |
FAM72A |
family with sequence similarity 72, member A |
| chr3_+_137906154 | 0.07 |
ENST00000466749.1 ENST00000358441.2 ENST00000489213.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr1_-_143913143 | 0.07 |
ENST00000400889.1 |
FAM72D |
family with sequence similarity 72, member D |
| chr3_+_47021168 | 0.07 |
ENST00000450053.3 ENST00000292309.5 ENST00000383740.2 |
NBEAL2 |
neurobeachin-like 2 |
| chr15_+_49447947 | 0.07 |
ENST00000327171.3 ENST00000560654.1 |
GALK2 |
galactokinase 2 |
| chr14_+_77787227 | 0.07 |
ENST00000216465.5 ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1 |
glutathione S-transferase zeta 1 |
| chr12_-_52967600 | 0.07 |
ENST00000549343.1 ENST00000305620.2 |
KRT74 |
keratin 74 |
| chr2_-_242576864 | 0.07 |
ENST00000407315.1 |
THAP4 |
THAP domain containing 4 |
| chr11_-_6704513 | 0.07 |
ENST00000532203.1 ENST00000288937.6 |
MRPL17 |
mitochondrial ribosomal protein L17 |
| chr19_-_42806842 | 0.07 |
ENST00000596265.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr1_+_235492300 | 0.07 |
ENST00000476121.1 ENST00000497327.1 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
| chr3_+_50654550 | 0.07 |
ENST00000430409.1 ENST00000357955.2 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
| chr22_+_29469012 | 0.07 |
ENST00000400335.4 ENST00000400338.2 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr2_-_54197915 | 0.07 |
ENST00000404125.1 |
PSME4 |
proteasome (prosome, macropain) activator subunit 4 |
| chr1_-_74663825 | 0.06 |
ENST00000370911.3 ENST00000370909.2 ENST00000354431.4 |
LRRIQ3 |
leucine-rich repeats and IQ motif containing 3 |
| chr1_+_235490659 | 0.06 |
ENST00000488594.1 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
| chr21_+_17102311 | 0.06 |
ENST00000285679.6 ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25 |
ubiquitin specific peptidase 25 |
| chr19_-_49496557 | 0.06 |
ENST00000323798.3 ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1 |
glycogen synthase 1 (muscle) |
| chr19_-_10444188 | 0.06 |
ENST00000293677.6 |
RAVER1 |
ribonucleoprotein, PTB-binding 1 |
| chr14_+_54863739 | 0.06 |
ENST00000541304.1 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr13_-_46961317 | 0.06 |
ENST00000322896.6 |
KIAA0226L |
KIAA0226-like |
| chr1_-_115300592 | 0.06 |
ENST00000261443.5 ENST00000534699.1 ENST00000339438.6 ENST00000529046.1 ENST00000525970.1 ENST00000369530.1 ENST00000530886.1 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chr11_-_61684962 | 0.06 |
ENST00000394836.2 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
| chr22_-_20104700 | 0.06 |
ENST00000439169.2 ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A |
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr17_-_41132410 | 0.06 |
ENST00000409446.3 ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr15_+_41221536 | 0.06 |
ENST00000249749.5 |
DLL4 |
delta-like 4 (Drosophila) |
| chr19_-_19030157 | 0.06 |
ENST00000349893.4 ENST00000351079.4 ENST00000600932.1 ENST00000262812.4 |
COPE |
coatomer protein complex, subunit epsilon |
| chr11_+_124735282 | 0.06 |
ENST00000397801.1 |
ROBO3 |
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr5_+_154238042 | 0.06 |
ENST00000519211.1 ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr17_-_42277203 | 0.06 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr19_-_14640005 | 0.06 |
ENST00000596853.1 ENST00000596075.1 ENST00000595992.1 ENST00000396969.4 ENST00000601533.1 ENST00000598692.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr17_-_7387524 | 0.06 |
ENST00000311403.4 |
ZBTB4 |
zinc finger and BTB domain containing 4 |
| chr1_+_183441600 | 0.06 |
ENST00000367537.3 |
SMG7 |
SMG7 nonsense mediated mRNA decay factor |
| chr15_-_99791413 | 0.06 |
ENST00000394129.2 ENST00000558663.1 ENST00000394135.3 ENST00000561365.1 ENST00000560279.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr2_+_130939235 | 0.06 |
ENST00000425361.1 ENST00000457492.1 |
MZT2B |
mitotic spindle organizing protein 2B |
| chr14_+_76452090 | 0.06 |
ENST00000314067.6 ENST00000238628.6 ENST00000556742.1 |
IFT43 |
intraflagellar transport 43 homolog (Chlamydomonas) |
| chr3_-_49395705 | 0.05 |
ENST00000419349.1 |
GPX1 |
glutathione peroxidase 1 |
| chr1_+_214454492 | 0.05 |
ENST00000366957.5 ENST00000415093.2 |
SMYD2 |
SET and MYND domain containing 2 |
| chr11_+_125774258 | 0.05 |
ENST00000263576.6 |
DDX25 |
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr14_+_54863682 | 0.05 |
ENST00000543789.2 ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr7_+_100271446 | 0.05 |
ENST00000419828.1 ENST00000427895.1 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr14_+_54863667 | 0.05 |
ENST00000335183.6 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr2_+_135809835 | 0.05 |
ENST00000264158.8 ENST00000539493.1 ENST00000442034.1 ENST00000425393.1 |
RAB3GAP1 |
RAB3 GTPase activating protein subunit 1 (catalytic) |
| chr11_+_125774362 | 0.05 |
ENST00000530414.1 ENST00000530129.2 |
DDX25 |
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr4_+_164415594 | 0.05 |
ENST00000509657.1 ENST00000358572.5 |
TMA16 |
translation machinery associated 16 homolog (S. cerevisiae) |
| chr1_-_40237020 | 0.05 |
ENST00000327582.5 |
OXCT2 |
3-oxoacid CoA transferase 2 |
| chr9_+_132597722 | 0.05 |
ENST00000372429.3 ENST00000315480.4 ENST00000358355.1 |
USP20 |
ubiquitin specific peptidase 20 |
| chr17_+_7155556 | 0.05 |
ENST00000570500.1 ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr17_+_3627185 | 0.05 |
ENST00000325418.4 |
GSG2 |
germ cell associated 2 (haspin) |
| chr3_-_49395892 | 0.05 |
ENST00000419783.1 |
GPX1 |
glutathione peroxidase 1 |
| chr9_+_114393634 | 0.05 |
ENST00000556107.1 ENST00000374294.3 |
DNAJC25 DNAJC25-GNG10 |
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr17_+_40913210 | 0.05 |
ENST00000253796.5 |
RAMP2 |
receptor (G protein-coupled) activity modifying protein 2 |
| chr1_+_7831323 | 0.05 |
ENST00000054666.6 |
VAMP3 |
vesicle-associated membrane protein 3 |
| chr11_+_65554493 | 0.05 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr5_+_154237778 | 0.05 |
ENST00000523698.1 ENST00000517876.1 ENST00000520472.1 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr14_-_92588246 | 0.05 |
ENST00000329559.3 |
NDUFB1 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr7_+_100271355 | 0.05 |
ENST00000436220.1 ENST00000424361.1 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr17_+_6915730 | 0.05 |
ENST00000548577.1 |
RNASEK |
ribonuclease, RNase K |
| chr5_+_154238096 | 0.05 |
ENST00000517568.1 ENST00000524105.1 ENST00000285896.6 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr11_+_1860200 | 0.05 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr19_+_33463127 | 0.05 |
ENST00000589646.1 ENST00000588258.1 ENST00000590179.1 |
C19orf40 |
chromosome 19 open reading frame 40 |
| chr17_+_62075703 | 0.05 |
ENST00000577953.1 ENST00000582540.1 ENST00000579184.1 ENST00000425164.3 ENST00000412177.1 ENST00000539996.1 ENST00000583891.1 ENST00000580752.1 |
C17orf72 |
chromosome 17 open reading frame 72 |
| chr14_+_92588281 | 0.05 |
ENST00000298875.4 ENST00000553427.1 |
CPSF2 |
cleavage and polyadenylation specific factor 2, 100kDa |
| chr19_-_48867291 | 0.05 |
ENST00000435956.3 |
TMEM143 |
transmembrane protein 143 |
| chr22_+_39916558 | 0.05 |
ENST00000337304.2 ENST00000396680.1 |
ATF4 |
activating transcription factor 4 |
| chr16_-_57831676 | 0.05 |
ENST00000465878.2 ENST00000539578.1 ENST00000561524.1 |
KIFC3 |
kinesin family member C3 |
| chr17_+_6915798 | 0.05 |
ENST00000402093.1 |
RNASEK |
ribonuclease, RNase K |
| chr17_+_7387919 | 0.05 |
ENST00000572844.1 |
POLR2A |
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr4_-_103747011 | 0.04 |
ENST00000350435.7 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr5_+_76506706 | 0.04 |
ENST00000340978.3 ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B |
phosphodiesterase 8B |
| chr19_-_48867171 | 0.04 |
ENST00000377431.2 ENST00000436660.2 ENST00000541566.1 |
TMEM143 |
transmembrane protein 143 |
| chr14_-_92588013 | 0.04 |
ENST00000553514.1 ENST00000605997.1 |
NDUFB1 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr13_+_27131887 | 0.04 |
ENST00000335327.5 |
WASF3 |
WAS protein family, member 3 |
| chrX_+_16804544 | 0.04 |
ENST00000380122.5 ENST00000398155.4 |
TXLNG |
taxilin gamma |
| chr7_+_131012605 | 0.04 |
ENST00000446815.1 ENST00000352689.6 |
MKLN1 |
muskelin 1, intracellular mediator containing kelch motifs |
| chrX_+_38660682 | 0.04 |
ENST00000457894.1 |
MID1IP1 |
MID1 interacting protein 1 |
| chr11_+_73882311 | 0.04 |
ENST00000398427.4 ENST00000544401.1 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr9_+_37422663 | 0.04 |
ENST00000318158.6 ENST00000607784.1 |
GRHPR |
glyoxylate reductase/hydroxypyruvate reductase |
| chr11_+_73882144 | 0.04 |
ENST00000328257.8 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr10_-_99205607 | 0.04 |
ENST00000477692.2 ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1 |
exosome component 1 |
| chr12_-_54071181 | 0.04 |
ENST00000338662.5 |
ATP5G2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr15_+_66161792 | 0.04 |
ENST00000564910.1 ENST00000261890.2 |
RAB11A |
RAB11A, member RAS oncogene family |
| chr7_+_5111723 | 0.04 |
ENST00000498308.1 |
RBAKDN |
RBAK downstream neighbor (non-protein coding) |
| chr12_+_56109926 | 0.04 |
ENST00000547076.1 |
BLOC1S1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr19_-_34012674 | 0.04 |
ENST00000436370.3 ENST00000397032.4 ENST00000244137.7 |
PEPD |
peptidase D |
| chr7_+_138916231 | 0.04 |
ENST00000473989.3 ENST00000288561.8 |
UBN2 |
ubinuclein 2 |
| chr8_-_125551278 | 0.04 |
ENST00000519232.1 ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1 |
TatD DNase domain containing 1 |
| chr2_+_153191706 | 0.04 |
ENST00000288670.9 |
FMNL2 |
formin-like 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.2 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.2 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.1 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.0 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.2 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |