Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
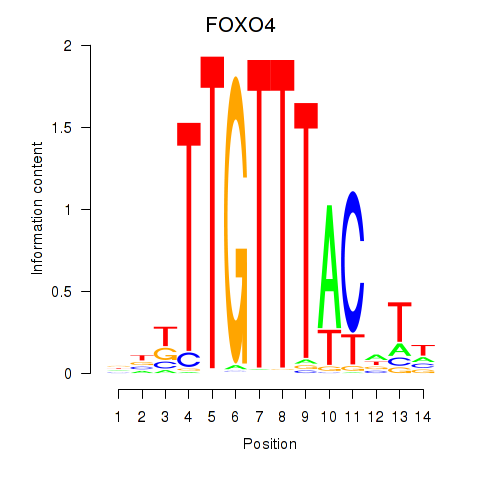
Results for FOXO4
Z-value: 0.94
Transcription factors associated with FOXO4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO4
|
ENSG00000184481.12 | FOXO4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO4 | hg19_v2_chrX_+_70316005_70316047 | -0.58 | 1.3e-01 | Click! |
Activity profile of FOXO4 motif
Sorted Z-values of FOXO4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_57045228 | 2.21 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr9_-_95186739 | 1.43 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr6_-_152489484 | 1.26 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr10_+_70847852 | 1.26 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr14_-_60097297 | 1.22 |
ENST00000395090.1 |
RTN1 |
reticulon 1 |
| chr14_-_61124977 | 1.08 |
ENST00000554986.1 |
SIX1 |
SIX homeobox 1 |
| chr4_-_70626314 | 1.05 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr5_+_156712372 | 0.94 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr4_-_70626430 | 0.85 |
ENST00000310613.3 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr5_-_137674000 | 0.80 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr8_-_13134045 | 0.75 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr6_+_72596604 | 0.74 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr3_+_148447887 | 0.63 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr4_+_124320665 | 0.63 |
ENST00000394339.2 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chrX_+_9431324 | 0.62 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr1_+_145524891 | 0.61 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr17_-_67138015 | 0.58 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr6_-_117747015 | 0.57 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr21_+_37507210 | 0.57 |
ENST00000290354.5 |
CBR3 |
carbonyl reductase 3 |
| chr4_-_159080806 | 0.56 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr7_+_134551583 | 0.50 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr12_-_92539614 | 0.50 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr10_-_25241499 | 0.49 |
ENST00000376378.1 ENST00000376376.3 ENST00000320152.6 |
PRTFDC1 |
phosphoribosyl transferase domain containing 1 |
| chr6_+_108882069 | 0.48 |
ENST00000406360.1 |
FOXO3 |
forkhead box O3 |
| chr2_+_109204743 | 0.46 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr13_+_32838801 | 0.45 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chr8_+_70404996 | 0.44 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr15_-_52970820 | 0.44 |
ENST00000261844.7 ENST00000399202.4 ENST00000562135.1 |
FAM214A |
family with sequence similarity 214, member A |
| chr6_-_31651817 | 0.42 |
ENST00000375863.3 ENST00000375860.2 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr5_-_64920115 | 0.42 |
ENST00000381018.3 ENST00000274327.7 |
TRIM23 |
tripartite motif containing 23 |
| chr2_-_192711968 | 0.42 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr11_+_77532155 | 0.42 |
ENST00000532481.1 ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr11_-_89224139 | 0.41 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_89223883 | 0.41 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr18_+_6834472 | 0.40 |
ENST00000581099.1 ENST00000419673.2 ENST00000531294.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr7_-_14028488 | 0.39 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr11_+_77532233 | 0.39 |
ENST00000525409.1 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr7_+_120628731 | 0.39 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr1_+_97188188 | 0.39 |
ENST00000541987.1 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr1_+_150122034 | 0.38 |
ENST00000025469.6 ENST00000369124.4 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
| chr11_+_12115543 | 0.38 |
ENST00000537344.1 ENST00000532179.1 ENST00000526065.1 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr2_-_101767715 | 0.37 |
ENST00000376840.4 ENST00000409318.1 |
TBC1D8 |
TBC1 domain family, member 8 (with GRAM domain) |
| chr2_+_175260514 | 0.35 |
ENST00000424069.1 ENST00000427038.1 |
SCRN3 |
secernin 3 |
| chr6_-_87804815 | 0.35 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr1_-_182360498 | 0.35 |
ENST00000417584.2 |
GLUL |
glutamate-ammonia ligase |
| chr4_-_186696425 | 0.35 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr5_+_140529630 | 0.34 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chr10_-_90751038 | 0.33 |
ENST00000458159.1 ENST00000415557.1 ENST00000458208.1 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr20_+_45338126 | 0.33 |
ENST00000359271.2 |
SLC2A10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr2_+_175260451 | 0.33 |
ENST00000458563.1 ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3 |
secernin 3 |
| chr20_+_18125727 | 0.33 |
ENST00000489634.2 |
CSRP2BP |
CSRP2 binding protein |
| chr10_-_25010795 | 0.32 |
ENST00000416305.1 ENST00000376410.2 |
ARHGAP21 |
Rho GTPase activating protein 21 |
| chr15_-_55657428 | 0.32 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr2_+_233527443 | 0.31 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr6_-_13621126 | 0.31 |
ENST00000600057.1 |
AL441883.1 |
Uncharacterized protein |
| chr1_-_182360918 | 0.31 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chr17_-_8263538 | 0.30 |
ENST00000535173.1 |
AC135178.1 |
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr7_+_151038850 | 0.29 |
ENST00000355851.4 ENST00000566856.1 ENST00000470229.1 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
| chr6_-_167571817 | 0.29 |
ENST00000366834.1 |
GPR31 |
G protein-coupled receptor 31 |
| chr9_+_108006880 | 0.28 |
ENST00000374723.1 ENST00000374720.3 ENST00000374724.1 |
SLC44A1 |
solute carrier family 44 (choline transporter), member 1 |
| chr10_-_4285923 | 0.28 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr9_+_5510558 | 0.28 |
ENST00000397747.3 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
| chr3_-_71632894 | 0.28 |
ENST00000493089.1 |
FOXP1 |
forkhead box P1 |
| chr1_-_227505289 | 0.27 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr12_-_7596735 | 0.27 |
ENST00000416109.2 ENST00000396630.1 ENST00000313599.3 |
CD163L1 |
CD163 molecule-like 1 |
| chr2_-_166060571 | 0.27 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr19_+_18111927 | 0.27 |
ENST00000379656.3 |
ARRDC2 |
arrestin domain containing 2 |
| chr10_-_49482907 | 0.26 |
ENST00000374201.3 ENST00000407470.4 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr9_+_5510492 | 0.26 |
ENST00000397745.2 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
| chr5_-_131132658 | 0.26 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr7_+_7811992 | 0.25 |
ENST00000406829.1 |
RPA3-AS1 |
RPA3 antisense RNA 1 |
| chr12_+_100041527 | 0.25 |
ENST00000324341.1 |
FAM71C |
family with sequence similarity 71, member C |
| chr6_+_101846664 | 0.25 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr2_-_166060552 | 0.25 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr3_-_18466026 | 0.25 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr4_-_100242549 | 0.24 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr5_+_137203465 | 0.24 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr3_-_185826855 | 0.24 |
ENST00000306376.5 |
ETV5 |
ets variant 5 |
| chr1_-_241803679 | 0.24 |
ENST00000331838.5 |
OPN3 |
opsin 3 |
| chr1_-_241803649 | 0.23 |
ENST00000366554.2 |
OPN3 |
opsin 3 |
| chr19_-_58662139 | 0.23 |
ENST00000598312.1 |
ZNF329 |
zinc finger protein 329 |
| chr11_+_33061543 | 0.23 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr2_+_109204909 | 0.22 |
ENST00000393310.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr16_-_15736881 | 0.22 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr2_+_12857015 | 0.22 |
ENST00000155926.4 |
TRIB2 |
tribbles pseudokinase 2 |
| chr2_+_65283506 | 0.22 |
ENST00000377990.2 |
CEP68 |
centrosomal protein 68kDa |
| chr9_-_86432547 | 0.22 |
ENST00000376365.3 ENST00000376371.2 |
GKAP1 |
G kinase anchoring protein 1 |
| chr2_+_12857043 | 0.22 |
ENST00000381465.2 |
TRIB2 |
tribbles pseudokinase 2 |
| chr1_-_54405773 | 0.22 |
ENST00000371376.1 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr17_-_30185946 | 0.22 |
ENST00000579741.1 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
| chr1_+_174844645 | 0.21 |
ENST00000486220.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr19_-_40732594 | 0.21 |
ENST00000430325.2 ENST00000433940.1 |
CNTD2 |
cyclin N-terminal domain containing 2 |
| chr14_+_23067146 | 0.21 |
ENST00000428304.2 |
ABHD4 |
abhydrolase domain containing 4 |
| chr19_+_50380682 | 0.21 |
ENST00000221543.5 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr13_+_24144509 | 0.21 |
ENST00000248484.4 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr5_-_125930929 | 0.21 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr11_-_77531752 | 0.21 |
ENST00000440064.2 ENST00000528095.1 |
RSF1 |
remodeling and spacing factor 1 |
| chr19_+_50380917 | 0.21 |
ENST00000535102.2 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr5_-_133510456 | 0.20 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr20_+_17680587 | 0.20 |
ENST00000427254.1 ENST00000377805.3 |
BANF2 |
barrier to autointegration factor 2 |
| chr1_-_47655686 | 0.20 |
ENST00000294338.2 |
PDZK1IP1 |
PDZK1 interacting protein 1 |
| chr17_+_44668035 | 0.20 |
ENST00000398238.4 ENST00000225282.8 |
NSF |
N-ethylmaleimide-sensitive factor |
| chr2_+_65283529 | 0.19 |
ENST00000546106.1 ENST00000537589.1 ENST00000260569.4 |
CEP68 |
centrosomal protein 68kDa |
| chr2_+_201980827 | 0.19 |
ENST00000309955.3 ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr17_+_33474860 | 0.19 |
ENST00000394570.2 |
UNC45B |
unc-45 homolog B (C. elegans) |
| chr17_+_33474826 | 0.19 |
ENST00000268876.5 ENST00000433649.1 ENST00000378449.1 |
UNC45B |
unc-45 homolog B (C. elegans) |
| chr5_+_98104978 | 0.18 |
ENST00000308234.7 |
RGMB |
repulsive guidance molecule family member b |
| chr11_-_89224299 | 0.18 |
ENST00000343727.5 ENST00000531342.1 ENST00000375979.3 |
NOX4 |
NADPH oxidase 4 |
| chr17_-_30185971 | 0.18 |
ENST00000378634.2 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
| chr18_+_72922710 | 0.18 |
ENST00000322038.5 |
TSHZ1 |
teashirt zinc finger homeobox 1 |
| chr4_-_152149033 | 0.18 |
ENST00000514152.1 |
SH3D19 |
SH3 domain containing 19 |
| chr7_-_38394118 | 0.18 |
ENST00000390345.2 |
TRGV4 |
T cell receptor gamma variable 4 |
| chr1_-_43855479 | 0.18 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr17_+_79953310 | 0.18 |
ENST00000582355.2 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr5_+_135468516 | 0.17 |
ENST00000507118.1 ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5 |
SMAD family member 5 |
| chr7_-_6098770 | 0.17 |
ENST00000536084.1 ENST00000446699.1 ENST00000199389.6 |
EIF2AK1 |
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr22_+_45809560 | 0.17 |
ENST00000342894.3 ENST00000538017.1 |
RIBC2 |
RIB43A domain with coiled-coils 2 |
| chr8_-_57472154 | 0.17 |
ENST00000499425.1 ENST00000523664.1 ENST00000518943.1 ENST00000524338.1 |
LINC00968 |
long intergenic non-protein coding RNA 968 |
| chr1_-_207095212 | 0.17 |
ENST00000420007.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr7_+_134832808 | 0.17 |
ENST00000275767.3 |
TMEM140 |
transmembrane protein 140 |
| chr5_-_42811986 | 0.17 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr19_-_7293942 | 0.17 |
ENST00000341500.5 ENST00000302850.5 |
INSR |
insulin receptor |
| chr1_-_157811588 | 0.17 |
ENST00000368174.4 |
CD5L |
CD5 molecule-like |
| chr4_+_88754113 | 0.17 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr3_+_101504200 | 0.17 |
ENST00000422132.1 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr1_-_227505826 | 0.16 |
ENST00000334218.5 ENST00000366766.2 ENST00000366764.2 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr5_-_179047881 | 0.16 |
ENST00000521173.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr6_+_21666633 | 0.16 |
ENST00000606851.1 |
CASC15 |
cancer susceptibility candidate 15 (non-protein coding) |
| chrX_+_100743031 | 0.16 |
ENST00000423738.3 |
ARMCX4 |
armadillo repeat containing, X-linked 4 |
| chr19_+_30863271 | 0.15 |
ENST00000355537.3 |
ZNF536 |
zinc finger protein 536 |
| chr6_-_42016385 | 0.15 |
ENST00000502771.1 ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3 |
cyclin D3 |
| chr9_-_15472730 | 0.15 |
ENST00000481862.1 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr11_-_55703876 | 0.15 |
ENST00000301532.3 |
OR5I1 |
olfactory receptor, family 5, subfamily I, member 1 |
| chr8_-_62602327 | 0.15 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr7_-_144435985 | 0.14 |
ENST00000549981.1 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr15_-_40600026 | 0.14 |
ENST00000456256.2 ENST00000557821.1 |
PLCB2 |
phospholipase C, beta 2 |
| chr1_+_10490127 | 0.14 |
ENST00000602787.1 ENST00000602296.1 ENST00000400900.2 |
APITD1 APITD1-CORT |
apoptosis-inducing, TAF9-like domain 1 APITD1-CORT readthrough |
| chr3_-_71294304 | 0.14 |
ENST00000498215.1 |
FOXP1 |
forkhead box P1 |
| chr13_-_39564993 | 0.14 |
ENST00000423210.1 |
STOML3 |
stomatin (EPB72)-like 3 |
| chrX_+_120181457 | 0.14 |
ENST00000328078.1 |
GLUD2 |
glutamate dehydrogenase 2 |
| chr1_-_159684371 | 0.14 |
ENST00000255030.5 ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP |
C-reactive protein, pentraxin-related |
| chr12_+_6881678 | 0.14 |
ENST00000441671.2 ENST00000203629.2 |
LAG3 |
lymphocyte-activation gene 3 |
| chrX_+_106045891 | 0.14 |
ENST00000357242.5 ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
| chr11_-_77531858 | 0.13 |
ENST00000360355.2 |
RSF1 |
remodeling and spacing factor 1 |
| chr13_+_98086445 | 0.13 |
ENST00000245304.4 |
RAP2A |
RAP2A, member of RAS oncogene family |
| chr12_-_102591604 | 0.13 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr4_+_88754069 | 0.13 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr5_+_133706865 | 0.13 |
ENST00000265339.2 |
UBE2B |
ubiquitin-conjugating enzyme E2B |
| chr1_-_20126365 | 0.13 |
ENST00000294543.6 ENST00000375122.2 |
TMCO4 |
transmembrane and coiled-coil domains 4 |
| chr9_-_13175823 | 0.13 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr12_-_772901 | 0.13 |
ENST00000305108.4 |
NINJ2 |
ninjurin 2 |
| chr7_-_95064264 | 0.13 |
ENST00000536183.1 ENST00000433091.2 ENST00000222572.3 |
PON2 |
paraoxonase 2 |
| chr3_-_141868357 | 0.13 |
ENST00000489671.1 ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_121416437 | 0.13 |
ENST00000402929.1 ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A |
HNF1 homeobox A |
| chr13_+_24144796 | 0.13 |
ENST00000403372.2 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_226265364 | 0.13 |
ENST00000272907.6 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr5_-_107703556 | 0.13 |
ENST00000496714.1 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
| chr1_+_63989004 | 0.12 |
ENST00000371088.4 |
EFCAB7 |
EF-hand calcium binding domain 7 |
| chr1_+_236686717 | 0.12 |
ENST00000341872.6 ENST00000450372.2 |
LGALS8 |
lectin, galactoside-binding, soluble, 8 |
| chr11_+_35684288 | 0.12 |
ENST00000299413.5 |
TRIM44 |
tripartite motif containing 44 |
| chr13_+_103459704 | 0.12 |
ENST00000602836.1 |
BIVM-ERCC5 |
BIVM-ERCC5 readthrough |
| chr10_-_13570533 | 0.12 |
ENST00000396900.2 ENST00000396898.2 |
BEND7 |
BEN domain containing 7 |
| chr10_-_82049424 | 0.12 |
ENST00000372213.3 |
MAT1A |
methionine adenosyltransferase I, alpha |
| chr2_+_170683942 | 0.12 |
ENST00000272793.5 |
UBR3 |
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr18_+_32073253 | 0.12 |
ENST00000283365.9 ENST00000596745.1 ENST00000315456.6 |
DTNA |
dystrobrevin, alpha |
| chr11_-_82745238 | 0.12 |
ENST00000531021.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr20_-_62582475 | 0.12 |
ENST00000369908.5 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
| chr18_-_30716038 | 0.12 |
ENST00000581852.1 |
CCDC178 |
coiled-coil domain containing 178 |
| chr1_-_102312517 | 0.12 |
ENST00000338858.5 |
OLFM3 |
olfactomedin 3 |
| chr17_+_6659153 | 0.11 |
ENST00000441631.1 ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1 |
XIAP associated factor 1 |
| chrX_-_131623874 | 0.11 |
ENST00000436215.1 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr2_+_149402553 | 0.11 |
ENST00000258484.6 ENST00000409654.1 |
EPC2 |
enhancer of polycomb homolog 2 (Drosophila) |
| chr6_+_126277842 | 0.11 |
ENST00000229633.5 |
HINT3 |
histidine triad nucleotide binding protein 3 |
| chr1_+_43855560 | 0.11 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr19_+_13906250 | 0.11 |
ENST00000254323.2 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
| chr8_-_42623747 | 0.11 |
ENST00000534622.1 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr3_-_28390581 | 0.11 |
ENST00000479665.1 |
AZI2 |
5-azacytidine induced 2 |
| chr2_+_170683979 | 0.11 |
ENST00000418381.1 |
UBR3 |
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr8_-_42623924 | 0.11 |
ENST00000276410.2 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr17_-_40897043 | 0.11 |
ENST00000428826.2 ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1 |
enhancer of zeste homolog 1 (Drosophila) |
| chr7_+_12250886 | 0.11 |
ENST00000444443.1 ENST00000396667.3 |
TMEM106B |
transmembrane protein 106B |
| chr8_+_49984894 | 0.10 |
ENST00000522267.1 ENST00000399653.4 ENST00000303202.8 |
C8orf22 |
chromosome 8 open reading frame 22 |
| chr12_-_100486668 | 0.10 |
ENST00000550544.1 ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L |
UHRF1 binding protein 1-like |
| chr3_-_150920979 | 0.10 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr7_-_149158187 | 0.10 |
ENST00000247930.4 |
ZNF777 |
zinc finger protein 777 |
| chr11_-_10879572 | 0.10 |
ENST00000413761.2 |
ZBED5 |
zinc finger, BED-type containing 5 |
| chr4_+_78804393 | 0.10 |
ENST00000502384.1 |
MRPL1 |
mitochondrial ribosomal protein L1 |
| chr19_+_859654 | 0.10 |
ENST00000592860.1 |
CFD |
complement factor D (adipsin) |
| chr11_-_10879593 | 0.10 |
ENST00000528289.1 ENST00000432999.2 |
ZBED5 |
zinc finger, BED-type containing 5 |
| chr11_-_77532050 | 0.10 |
ENST00000308488.6 |
RSF1 |
remodeling and spacing factor 1 |
| chr12_+_96588143 | 0.10 |
ENST00000228741.3 ENST00000547249.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr16_+_1730338 | 0.10 |
ENST00000566691.1 ENST00000382710.4 |
HN1L |
hematological and neurological expressed 1-like |
| chrX_-_19689106 | 0.10 |
ENST00000379716.1 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr12_+_54378923 | 0.09 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr16_-_18887627 | 0.09 |
ENST00000563235.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr12_-_68696652 | 0.09 |
ENST00000539972.1 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
| chr3_+_107096188 | 0.09 |
ENST00000261058.1 |
CCDC54 |
coiled-coil domain containing 54 |
| chr1_+_162039558 | 0.09 |
ENST00000530878.1 ENST00000361897.5 |
NOS1AP |
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr6_+_168418553 | 0.09 |
ENST00000354419.2 ENST00000351261.3 |
KIF25 |
kinesin family member 25 |
| chr19_-_50380536 | 0.09 |
ENST00000391832.3 ENST00000391834.2 ENST00000344175.5 |
AKT1S1 |
AKT1 substrate 1 (proline-rich) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.3 | 1.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.6 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 0.5 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 0.5 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.2 | 0.6 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 1.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.6 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.5 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.4 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 1.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.2 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.1 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.8 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.2 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.0 | 0.7 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:1905033 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 1.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.8 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0022027 | positive regulation of centrosome duplication(GO:0010825) interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.0 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 1.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.6 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 0.6 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.0 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |