Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
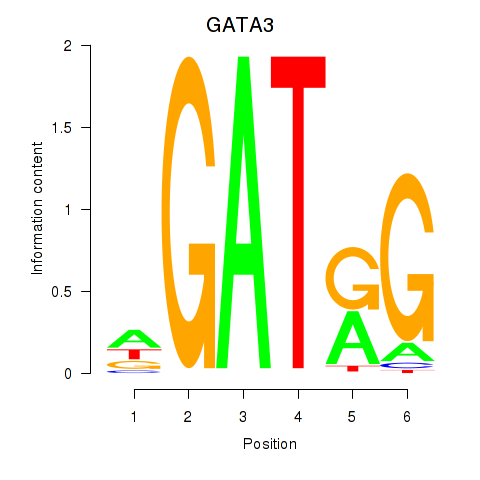
Results for GATA3
Z-value: 1.22
Transcription factors associated with GATA3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA3
|
ENSG00000107485.11 | GATA3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA3 | hg19_v2_chr10_+_8096631_8096660 | 0.58 | 1.3e-01 | Click! |
Activity profile of GATA3 motif
Sorted Z-values of GATA3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_30866564 | 0.90 |
ENST00000435069.1 ENST00000415957.2 ENST00000540910.1 |
SEC14L3 |
SEC14-like 3 (S. cerevisiae) |
| chr1_+_171060018 | 0.86 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr2_+_128177458 | 0.80 |
ENST00000409048.1 ENST00000422777.3 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr9_+_90112117 | 0.77 |
ENST00000358077.5 |
DAPK1 |
death-associated protein kinase 1 |
| chr8_-_93029865 | 0.76 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr16_+_55512742 | 0.75 |
ENST00000568715.1 ENST00000219070.4 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr11_-_35547151 | 0.72 |
ENST00000378878.3 ENST00000529303.1 ENST00000278360.3 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr3_-_121379739 | 0.69 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr20_+_12989596 | 0.68 |
ENST00000434210.1 ENST00000399002.2 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
| chr12_+_56075330 | 0.65 |
ENST00000394252.3 |
METTL7B |
methyltransferase like 7B |
| chr22_-_24641027 | 0.64 |
ENST00000398292.3 ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr6_-_31514516 | 0.63 |
ENST00000303892.5 ENST00000483251.1 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr15_-_93616340 | 0.60 |
ENST00000557420.1 ENST00000542321.2 |
RGMA |
repulsive guidance molecule family member a |
| chr8_-_93107443 | 0.58 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_+_49977818 | 0.57 |
ENST00000594009.1 ENST00000595510.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr15_-_93616892 | 0.57 |
ENST00000556658.1 ENST00000538818.1 ENST00000425933.2 |
RGMA |
repulsive guidance molecule family member a |
| chr4_-_138453606 | 0.57 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr12_-_91573132 | 0.54 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_-_91546926 | 0.54 |
ENST00000550758.1 |
DCN |
decorin |
| chr19_-_14945933 | 0.53 |
ENST00000322301.3 |
OR7A5 |
olfactory receptor, family 7, subfamily A, member 5 |
| chr3_+_45071622 | 0.52 |
ENST00000428034.1 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr8_-_93107696 | 0.51 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr11_-_111782484 | 0.51 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr1_+_78956651 | 0.51 |
ENST00000370757.3 ENST00000370756.3 |
PTGFR |
prostaglandin F receptor (FP) |
| chr3_-_114343768 | 0.50 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr11_-_35547572 | 0.50 |
ENST00000378880.2 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr4_-_159080806 | 0.49 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr4_+_55095264 | 0.49 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr1_+_114522049 | 0.49 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr16_+_29690358 | 0.49 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr19_+_47105309 | 0.49 |
ENST00000599839.1 ENST00000596362.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr2_+_128175997 | 0.48 |
ENST00000234071.3 ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr16_-_67260691 | 0.48 |
ENST00000447579.1 ENST00000393992.1 ENST00000424285.1 |
LRRC29 |
leucine rich repeat containing 29 |
| chr3_-_49170405 | 0.47 |
ENST00000305544.4 ENST00000494831.1 |
LAMB2 |
laminin, beta 2 (laminin S) |
| chr3_-_49170522 | 0.45 |
ENST00000418109.1 |
LAMB2 |
laminin, beta 2 (laminin S) |
| chr14_-_92413353 | 0.44 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr11_-_111781554 | 0.44 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr9_-_95244781 | 0.44 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr3_-_49722523 | 0.44 |
ENST00000448220.1 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr1_-_72748417 | 0.44 |
ENST00000357731.5 |
NEGR1 |
neuronal growth regulator 1 |
| chr11_+_117073850 | 0.44 |
ENST00000529622.1 |
TAGLN |
transgelin |
| chr1_-_1293904 | 0.44 |
ENST00000309212.6 ENST00000342753.4 ENST00000445648.2 |
MXRA8 |
matrix-remodelling associated 8 |
| chr17_-_76870222 | 0.43 |
ENST00000585421.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr17_-_53800217 | 0.43 |
ENST00000424486.2 |
TMEM100 |
transmembrane protein 100 |
| chr9_-_28026318 | 0.42 |
ENST00000308675.3 |
LINGO2 |
leucine rich repeat and Ig domain containing 2 |
| chr2_+_85646054 | 0.42 |
ENST00000389938.2 |
SH2D6 |
SH2 domain containing 6 |
| chr7_-_76255444 | 0.42 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr10_-_105212059 | 0.41 |
ENST00000260743.5 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr1_+_79086088 | 0.41 |
ENST00000370751.5 ENST00000342282.3 |
IFI44L |
interferon-induced protein 44-like |
| chr1_+_150245177 | 0.41 |
ENST00000369098.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr17_+_3100813 | 0.41 |
ENST00000381951.1 |
OR1A2 |
olfactory receptor, family 1, subfamily A, member 2 |
| chr1_-_156647189 | 0.41 |
ENST00000368223.3 |
NES |
nestin |
| chr6_+_72596604 | 0.41 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr13_+_102104980 | 0.41 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr8_-_93107827 | 0.40 |
ENST00000520724.1 ENST00000518844.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr12_-_54867352 | 0.40 |
ENST00000305879.5 |
GTSF1 |
gametocyte specific factor 1 |
| chr17_-_19290483 | 0.40 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr11_-_102401469 | 0.40 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr12_-_50298000 | 0.39 |
ENST00000550635.2 |
FAIM2 |
Fas apoptotic inhibitory molecule 2 |
| chr6_-_31514333 | 0.39 |
ENST00000376151.4 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr1_-_145470383 | 0.39 |
ENST00000369314.1 ENST00000369313.3 |
POLR3GL |
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like |
| chrX_-_1572629 | 0.39 |
ENST00000534940.1 |
ASMTL |
acetylserotonin O-methyltransferase-like |
| chr12_+_56862301 | 0.39 |
ENST00000338146.5 |
SPRYD4 |
SPRY domain containing 4 |
| chr17_-_76870126 | 0.39 |
ENST00000586057.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr9_-_115819039 | 0.38 |
ENST00000555206.1 |
ZFP37 |
ZFP37 zinc finger protein |
| chr9_+_125273081 | 0.38 |
ENST00000335302.5 |
OR1J2 |
olfactory receptor, family 1, subfamily J, member 2 |
| chr10_-_105212141 | 0.38 |
ENST00000369788.3 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr9_+_139871948 | 0.38 |
ENST00000224167.2 ENST00000457950.1 ENST00000371625.3 ENST00000371623.3 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr1_-_153517473 | 0.38 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr14_+_105939276 | 0.38 |
ENST00000483017.3 |
CRIP2 |
cysteine-rich protein 2 |
| chr4_+_62067860 | 0.37 |
ENST00000514591.1 |
LPHN3 |
latrophilin 3 |
| chr20_-_39928705 | 0.37 |
ENST00000436099.2 ENST00000309060.3 ENST00000373261.1 ENST00000436440.2 ENST00000540170.1 ENST00000557816.1 ENST00000560361.1 |
ZHX3 |
zinc fingers and homeoboxes 3 |
| chr9_+_124088860 | 0.37 |
ENST00000373806.1 |
GSN |
gelsolin |
| chr11_+_114166536 | 0.37 |
ENST00000299964.3 |
NNMT |
nicotinamide N-methyltransferase |
| chr9_-_21305312 | 0.37 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr17_-_19651668 | 0.36 |
ENST00000494157.2 ENST00000225740.6 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr21_+_27011899 | 0.36 |
ENST00000425221.2 |
JAM2 |
junctional adhesion molecule 2 |
| chr17_-_72864739 | 0.36 |
ENST00000579893.1 ENST00000544854.1 |
FDXR |
ferredoxin reductase |
| chr3_+_148447887 | 0.36 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr4_+_14113592 | 0.36 |
ENST00000502759.1 ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085 |
long intergenic non-protein coding RNA 1085 |
| chr13_+_93879085 | 0.36 |
ENST00000377047.4 |
GPC6 |
glypican 6 |
| chr20_+_34802295 | 0.36 |
ENST00000432603.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr1_+_145524891 | 0.36 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr12_-_91572278 | 0.36 |
ENST00000425043.1 ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN |
decorin |
| chr19_+_41117770 | 0.36 |
ENST00000601032.1 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr6_+_32812568 | 0.36 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr12_-_91573316 | 0.35 |
ENST00000393155.1 |
DCN |
decorin |
| chrX_+_135278908 | 0.35 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr11_+_5710919 | 0.35 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr1_-_46642154 | 0.35 |
ENST00000540385.1 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr4_-_186877502 | 0.35 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr4_-_140201333 | 0.35 |
ENST00000398955.1 |
MGARP |
mitochondria-localized glutamic acid-rich protein |
| chr13_+_102104952 | 0.35 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr3_+_148415571 | 0.34 |
ENST00000497524.1 ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr1_-_204116078 | 0.34 |
ENST00000367198.2 ENST00000452983.1 |
ETNK2 |
ethanolamine kinase 2 |
| chr10_-_127511790 | 0.34 |
ENST00000368797.4 ENST00000420761.1 |
UROS |
uroporphyrinogen III synthase |
| chr10_+_111967345 | 0.34 |
ENST00000332674.5 ENST00000453116.1 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr19_-_58864848 | 0.34 |
ENST00000263100.3 |
A1BG |
alpha-1-B glycoprotein |
| chr14_-_106406090 | 0.34 |
ENST00000390593.2 |
IGHV6-1 |
immunoglobulin heavy variable 6-1 |
| chr17_-_53809473 | 0.33 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr17_-_7590745 | 0.33 |
ENST00000514944.1 ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53 |
tumor protein p53 |
| chr19_+_16059818 | 0.33 |
ENST00000322107.1 |
OR10H4 |
olfactory receptor, family 10, subfamily H, member 4 |
| chr1_-_145039949 | 0.33 |
ENST00000313382.9 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr4_+_70916119 | 0.33 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chr11_-_65816591 | 0.33 |
ENST00000312006.4 |
GAL3ST3 |
galactose-3-O-sulfotransferase 3 |
| chr5_-_158526756 | 0.33 |
ENST00000313708.6 ENST00000517373.1 |
EBF1 |
early B-cell factor 1 |
| chr12_+_94656297 | 0.33 |
ENST00000545312.1 |
PLXNC1 |
plexin C1 |
| chr9_-_115818951 | 0.33 |
ENST00000553380.1 ENST00000374227.3 |
ZFP37 |
ZFP37 zinc finger protein |
| chr17_+_75447326 | 0.33 |
ENST00000591088.1 |
SEPT9 |
septin 9 |
| chr9_+_2029019 | 0.32 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_+_133320301 | 0.32 |
ENST00000352480.5 |
ASS1 |
argininosuccinate synthase 1 |
| chr7_+_134430212 | 0.32 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr17_-_4689649 | 0.32 |
ENST00000441199.2 ENST00000416307.2 |
VMO1 |
vitelline membrane outer layer 1 homolog (chicken) |
| chr2_+_189839046 | 0.32 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr2_+_24272576 | 0.32 |
ENST00000380986.4 ENST00000452109.1 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chrX_+_149531524 | 0.32 |
ENST00000370401.2 |
MAMLD1 |
mastermind-like domain containing 1 |
| chr4_+_113970772 | 0.32 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr5_+_32788945 | 0.32 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr8_-_19459993 | 0.32 |
ENST00000454498.2 ENST00000520003.1 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr22_-_31503490 | 0.32 |
ENST00000400299.2 |
SELM |
Selenoprotein M |
| chr7_+_142498725 | 0.32 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr6_+_69345166 | 0.31 |
ENST00000370598.1 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr19_-_50316489 | 0.31 |
ENST00000533418.1 |
FUZ |
fuzzy planar cell polarity protein |
| chr5_-_95158644 | 0.31 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr19_-_42894420 | 0.31 |
ENST00000597255.1 ENST00000222032.5 |
CNFN |
cornifelin |
| chr11_-_111782696 | 0.31 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr20_-_48099182 | 0.31 |
ENST00000371741.4 |
KCNB1 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr2_+_74685527 | 0.31 |
ENST00000393972.3 ENST00000409737.1 ENST00000428943.1 |
WBP1 |
WW domain binding protein 1 |
| chrX_+_135230712 | 0.31 |
ENST00000535737.1 |
FHL1 |
four and a half LIM domains 1 |
| chr4_-_186733363 | 0.31 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr10_-_81205373 | 0.30 |
ENST00000372336.3 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
| chr10_+_5135981 | 0.30 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr7_-_7782204 | 0.30 |
ENST00000418534.2 |
AC007161.5 |
AC007161.5 |
| chrX_+_152760397 | 0.30 |
ENST00000331595.4 ENST00000431891.1 |
BGN |
biglycan |
| chr19_+_47104553 | 0.30 |
ENST00000598871.1 ENST00000594523.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr1_+_163038565 | 0.30 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr5_-_95158375 | 0.30 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chr1_-_72566613 | 0.30 |
ENST00000306821.3 |
NEGR1 |
neuronal growth regulator 1 |
| chr1_-_109655355 | 0.30 |
ENST00000369945.3 |
C1orf194 |
chromosome 1 open reading frame 194 |
| chr19_-_48059113 | 0.30 |
ENST00000391901.3 ENST00000314121.4 ENST00000448976.1 |
ZNF541 |
zinc finger protein 541 |
| chr4_-_138453559 | 0.30 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr17_-_19651654 | 0.29 |
ENST00000395555.3 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr14_-_61124977 | 0.29 |
ENST00000554986.1 |
SIX1 |
SIX homeobox 1 |
| chr2_-_190044480 | 0.29 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr11_-_111781610 | 0.29 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr2_-_175869936 | 0.29 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr5_+_81575281 | 0.29 |
ENST00000380167.4 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr10_-_38265517 | 0.29 |
ENST00000302609.7 |
ZNF25 |
zinc finger protein 25 |
| chr19_-_36231437 | 0.29 |
ENST00000591748.1 |
IGFLR1 |
IGF-like family receptor 1 |
| chr11_-_111794446 | 0.29 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr17_+_36873677 | 0.28 |
ENST00000471200.1 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr12_+_94542459 | 0.28 |
ENST00000258526.4 |
PLXNC1 |
plexin C1 |
| chr19_+_10197463 | 0.28 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chrX_+_153455547 | 0.28 |
ENST00000430054.1 |
OPN1MW |
opsin 1 (cone pigments), medium-wave-sensitive |
| chr9_-_123239632 | 0.28 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr19_-_50316517 | 0.28 |
ENST00000313777.4 ENST00000445575.2 |
FUZ |
fuzzy planar cell polarity protein |
| chr9_+_130922537 | 0.28 |
ENST00000372994.1 |
C9orf16 |
chromosome 9 open reading frame 16 |
| chr16_+_20912075 | 0.28 |
ENST00000219168.4 |
LYRM1 |
LYR motif containing 1 |
| chr17_+_41158742 | 0.28 |
ENST00000415816.2 ENST00000438323.2 |
IFI35 |
interferon-induced protein 35 |
| chr12_-_54121261 | 0.28 |
ENST00000549784.1 ENST00000262059.4 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr3_-_49851313 | 0.28 |
ENST00000333486.3 |
UBA7 |
ubiquitin-like modifier activating enzyme 7 |
| chr11_+_124609742 | 0.28 |
ENST00000284292.6 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
| chr11_+_117070904 | 0.28 |
ENST00000529792.1 |
TAGLN |
transgelin |
| chr19_+_5681153 | 0.28 |
ENST00000579559.1 ENST00000577222.1 |
HSD11B1L RPL36 |
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr3_-_114343039 | 0.28 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr11_-_102826434 | 0.28 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr11_+_124609823 | 0.28 |
ENST00000412681.2 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
| chr17_-_19648916 | 0.27 |
ENST00000444455.1 ENST00000439102.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr10_-_4285923 | 0.27 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr15_+_55700741 | 0.27 |
ENST00000569691.1 |
C15orf65 |
chromosome 15 open reading frame 65 |
| chr1_-_153518270 | 0.27 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr11_-_105948040 | 0.27 |
ENST00000534815.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr16_-_1821496 | 0.27 |
ENST00000564628.1 ENST00000563498.1 |
NME3 |
NME/NM23 nucleoside diphosphate kinase 3 |
| chr19_-_50316423 | 0.27 |
ENST00000528094.1 ENST00000526575.1 |
FUZ |
fuzzy planar cell polarity protein |
| chr11_+_5372738 | 0.27 |
ENST00000380219.1 |
OR51B6 |
olfactory receptor, family 51, subfamily B, member 6 |
| chr1_-_55089191 | 0.27 |
ENST00000302250.2 ENST00000371304.2 |
FAM151A |
family with sequence similarity 151, member A |
| chr8_+_104383728 | 0.27 |
ENST00000330295.5 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr21_+_17961006 | 0.27 |
ENST00000602323.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr11_+_118215036 | 0.27 |
ENST00000392883.2 ENST00000532917.1 |
CD3G |
CD3g molecule, gamma (CD3-TCR complex) |
| chr16_-_28550348 | 0.27 |
ENST00000324873.6 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr16_-_3074231 | 0.27 |
ENST00000572355.1 ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1 |
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr19_-_51141196 | 0.27 |
ENST00000338916.4 |
SYT3 |
synaptotagmin III |
| chr10_+_112631547 | 0.26 |
ENST00000280154.7 ENST00000393104.2 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr11_-_57335750 | 0.26 |
ENST00000340573.4 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
| chr4_-_186732048 | 0.26 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr22_-_24622080 | 0.26 |
ENST00000425408.1 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr2_+_241807870 | 0.26 |
ENST00000307503.3 |
AGXT |
alanine-glyoxylate aminotransferase |
| chr2_-_211168332 | 0.26 |
ENST00000341685.4 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr9_+_133320339 | 0.26 |
ENST00000372394.1 ENST00000372393.3 ENST00000422569.1 |
ASS1 |
argininosuccinate synthase 1 |
| chr9_-_21217310 | 0.26 |
ENST00000380216.1 |
IFNA16 |
interferon, alpha 16 |
| chr14_+_21467414 | 0.26 |
ENST00000554422.1 ENST00000298681.4 |
SLC39A2 |
solute carrier family 39 (zinc transporter), member 2 |
| chr3_+_29322437 | 0.26 |
ENST00000434693.2 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chrX_+_103031421 | 0.26 |
ENST00000433491.1 ENST00000418604.1 ENST00000443502.1 |
PLP1 |
proteolipid protein 1 |
| chr4_-_186877806 | 0.26 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chrX_-_77395186 | 0.26 |
ENST00000341864.5 |
TAF9B |
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr6_-_52859046 | 0.26 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr7_-_38671098 | 0.26 |
ENST00000356264.2 |
AMPH |
amphiphysin |
| chr22_+_24577183 | 0.26 |
ENST00000358321.3 |
SUSD2 |
sushi domain containing 2 |
| chr10_+_111985713 | 0.26 |
ENST00000239007.7 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr5_+_180257951 | 0.26 |
ENST00000501855.2 |
LINC00847 |
long intergenic non-protein coding RNA 847 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 0.9 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.2 | 0.6 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 0.9 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.7 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 0.6 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.7 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.9 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.4 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.4 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.4 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.4 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.5 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.8 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.5 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.5 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.8 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.9 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 2.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.3 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.5 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.8 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.3 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.3 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.8 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.3 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.6 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.1 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.1 | 0.4 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 1.3 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 1.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.2 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 1.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.3 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.2 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.6 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.1 | 0.2 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 | 0.3 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.1 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.3 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.1 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.1 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.2 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.2 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.6 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.5 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.1 | 0.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.2 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.1 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 0.1 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.2 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) |
| 0.1 | 0.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.1 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.1 | 0.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.5 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.1 | GO:0051882 | mitochondrial depolarization(GO:0051882) |
| 0.1 | 0.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.9 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.2 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.6 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.2 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.2 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.1 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.2 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.0 | GO:0046031 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) ADP metabolic process(GO:0046031) |
| 0.0 | 0.1 | GO:1905154 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 1.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.3 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.4 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.1 | GO:1903818 | positive regulation of potassium ion transmembrane transporter activity(GO:1901018) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.2 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.4 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.6 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.2 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.8 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.2 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.0 | 1.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.0 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) |
| 0.0 | 0.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.0 | GO:1902744 | negative regulation of lamellipodium assembly(GO:0010593) negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032434) |
| 0.0 | 0.1 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.0 | 0.1 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.2 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.5 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.2 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 1.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0072019 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.0 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.0 | GO:0048705 | skeletal system morphogenesis(GO:0048705) |
| 0.0 | 0.1 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.0 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.0 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 1.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.4 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:1904172 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.2 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water deprivation(GO:0042631) cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.0 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.1 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0003068 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.3 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) |
| 0.0 | 0.1 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.8 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.0 | GO:1900225 | regulation of NLRP3 inflammasome complex assembly(GO:1900225) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.3 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.1 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.0 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.0 | GO:0034614 | cellular response to reactive oxygen species(GO:0034614) |
| 0.0 | 0.1 | GO:0071611 | granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) |
| 0.0 | 0.0 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.3 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.4 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 1.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.0 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.3 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.2 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:1903464 | negative regulation of mitotic cell cycle DNA replication(GO:1903464) |
| 0.0 | 0.3 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.0 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.4 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.0 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.1 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:1903507 | negative regulation of nucleic acid-templated transcription(GO:1903507) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 2.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.0 | GO:0016199 | axon choice point recognition(GO:0016198) axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.4 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 1.0 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.0 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) negative regulation of immunological synapse formation(GO:2000521) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.0 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.1 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.3 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.0 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.0 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.0 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 1.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:2001287 | regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.0 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.0 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.0 | 0.1 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.0 | GO:0021940 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:0072262 | metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.0 | GO:0042418 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.0 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.0 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.0 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 1.4 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.0 | 0.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.4 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.0 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.0 | GO:0032849 | regulation of cellular pH reduction(GO:0032847) positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.3 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.0 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.0 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.0 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.0 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.3 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.0 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.0 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0042992 | negative regulation of transcription factor import into nucleus(GO:0042992) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.2 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0046479 | glycosylceramide catabolic process(GO:0046477) glycosphingolipid catabolic process(GO:0046479) ceramide catabolic process(GO:0046514) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.0 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 2.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 0.8 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.6 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.5 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.6 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.3 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.6 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 1.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 1.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.1 | GO:0019867 | outer membrane(GO:0019867) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0099512 | supramolecular fiber(GO:0099512) polymeric cytoskeletal fiber(GO:0099513) |
| 0.0 | 0.0 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.5 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.2 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.0 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.1 | GO:0032994 | protein-lipid complex(GO:0032994) plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 2.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.0 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.7 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 0.8 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.2 | 0.5 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 0.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 0.5 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.4 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 0.4 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 1.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.7 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.4 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.4 | GO:0098639 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.3 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.4 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 1.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 1.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.3 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.3 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 1.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.2 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.1 | 0.7 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.3 | GO:0052841 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.3 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 1.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.6 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.2 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 1.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.3 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.5 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 4.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 2.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.0 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.0 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.0 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.0 | 0.0 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.0 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.0 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) amylin receptor activity(GO:0097643) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 4.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.7 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.0 | GO:0000988 | transcription factor activity, protein binding(GO:0000988) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0000828 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) inositol hexakisphosphate kinase activity(GO:0000828) inositol heptakisphosphate kinase activity(GO:0000829) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.0 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.1 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 2.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 3.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 1.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |