Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for GFI1
Z-value: 0.58
Transcription factors associated with GFI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1
|
ENSG00000162676.7 | GFI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1 | hg19_v2_chr1_-_92952433_92952489, hg19_v2_chr1_-_92951607_92951661, hg19_v2_chr1_-_92949505_92949543 | 0.80 | 1.7e-02 | Click! |
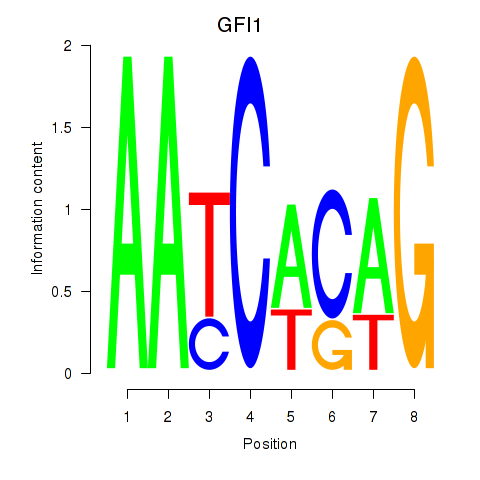
Activity profile of GFI1 motif
Sorted Z-values of GFI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21452804 | 1.06 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr18_+_21452964 | 1.05 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr1_-_242612779 | 0.91 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr20_+_58179582 | 0.69 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr8_-_57232656 | 0.63 |
ENST00000396721.2 |
SDR16C5 |
short chain dehydrogenase/reductase family 16C, member 5 |
| chr2_+_17721230 | 0.59 |
ENST00000457525.1 |
VSNL1 |
visinin-like 1 |
| chr18_+_29027696 | 0.58 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr19_-_10687907 | 0.56 |
ENST00000589348.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr5_+_36608422 | 0.55 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr2_+_17721920 | 0.49 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr11_-_5323226 | 0.48 |
ENST00000380224.1 |
OR51B4 |
olfactory receptor, family 51, subfamily B, member 4 |
| chr19_-_43969796 | 0.46 |
ENST00000244333.3 |
LYPD3 |
LY6/PLAUR domain containing 3 |
| chr12_+_44229846 | 0.45 |
ENST00000551577.1 ENST00000266534.3 |
TMEM117 |
transmembrane protein 117 |
| chr19_-_10687948 | 0.44 |
ENST00000592285.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr7_+_16793160 | 0.41 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr19_-_10687983 | 0.40 |
ENST00000587069.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr19_-_55660561 | 0.36 |
ENST00000587758.1 ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr14_+_21525981 | 0.33 |
ENST00000308227.2 |
RNASE8 |
ribonuclease, RNase A family, 8 |
| chr7_+_86273218 | 0.33 |
ENST00000361669.2 |
GRM3 |
glutamate receptor, metabotropic 3 |
| chr5_+_140261703 | 0.32 |
ENST00000409494.1 ENST00000289272.2 |
PCDHA13 |
protocadherin alpha 13 |
| chr13_+_113623509 | 0.31 |
ENST00000535094.2 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr1_+_62439037 | 0.30 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chrX_+_49832231 | 0.30 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr1_-_147232669 | 0.29 |
ENST00000369237.1 |
GJA5 |
gap junction protein, alpha 5, 40kDa |
| chr15_-_52587945 | 0.29 |
ENST00000443683.2 ENST00000558479.1 ENST00000261839.7 |
MYO5C |
myosin VC |
| chr2_+_85360499 | 0.29 |
ENST00000282111.3 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr17_-_56494882 | 0.29 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr17_-_56494713 | 0.29 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr19_-_16045220 | 0.29 |
ENST00000326742.8 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr17_-_56494908 | 0.29 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr17_-_39780819 | 0.28 |
ENST00000311208.8 |
KRT17 |
keratin 17 |
| chr4_+_41937131 | 0.28 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr19_-_16045665 | 0.28 |
ENST00000248041.8 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr14_-_22005062 | 0.27 |
ENST00000317492.5 |
SALL2 |
spalt-like transcription factor 2 |
| chr19_-_16045619 | 0.27 |
ENST00000402119.4 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr14_-_22005343 | 0.27 |
ENST00000327430.3 |
SALL2 |
spalt-like transcription factor 2 |
| chr12_+_56732658 | 0.26 |
ENST00000228534.4 |
IL23A |
interleukin 23, alpha subunit p19 |
| chr2_+_103378472 | 0.24 |
ENST00000412401.2 |
TMEM182 |
transmembrane protein 182 |
| chr10_-_10836865 | 0.24 |
ENST00000446372.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr16_+_4838412 | 0.24 |
ENST00000589327.1 |
SMIM22 |
small integral membrane protein 22 |
| chr8_-_145642267 | 0.24 |
ENST00000301305.3 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr19_-_43099070 | 0.23 |
ENST00000244336.5 |
CEACAM8 |
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr5_+_147582387 | 0.23 |
ENST00000325630.2 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr5_+_148206156 | 0.23 |
ENST00000305988.4 |
ADRB2 |
adrenoceptor beta 2, surface |
| chr3_-_33686743 | 0.23 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr1_+_156254070 | 0.23 |
ENST00000405535.2 ENST00000456810.1 |
TMEM79 |
transmembrane protein 79 |
| chr5_-_35230434 | 0.22 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr4_+_89299885 | 0.22 |
ENST00000380265.5 ENST00000273960.3 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr10_+_24498060 | 0.22 |
ENST00000376454.3 ENST00000376452.3 |
KIAA1217 |
KIAA1217 |
| chr16_+_4838393 | 0.22 |
ENST00000589721.1 |
SMIM22 |
small integral membrane protein 22 |
| chr10_+_24755416 | 0.22 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chr10_-_10836919 | 0.22 |
ENST00000602763.1 ENST00000415590.2 ENST00000434919.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr9_-_117150243 | 0.22 |
ENST00000374088.3 |
AKNA |
AT-hook transcription factor |
| chr4_+_89299994 | 0.22 |
ENST00000264346.7 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr5_-_78809950 | 0.21 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr18_-_78005231 | 0.21 |
ENST00000470488.2 ENST00000353265.3 |
PARD6G |
par-6 family cell polarity regulator gamma |
| chr1_-_25291475 | 0.21 |
ENST00000338888.3 ENST00000399916.1 |
RUNX3 |
runt-related transcription factor 3 |
| chr8_-_145641864 | 0.20 |
ENST00000276833.5 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr16_+_82090028 | 0.20 |
ENST00000568090.1 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chrX_-_133792480 | 0.20 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr14_+_71108460 | 0.20 |
ENST00000256367.2 |
TTC9 |
tetratricopeptide repeat domain 9 |
| chr15_+_92397051 | 0.20 |
ENST00000424469.2 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
| chr7_+_65670186 | 0.19 |
ENST00000304842.5 ENST00000442120.1 |
TPST1 |
tyrosylprotein sulfotransferase 1 |
| chr18_+_55711575 | 0.19 |
ENST00000356462.6 ENST00000400345.3 ENST00000589054.1 ENST00000256832.7 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_+_34642656 | 0.19 |
ENST00000257831.3 ENST00000450654.2 |
EHF |
ets homologous factor |
| chr1_-_10856694 | 0.19 |
ENST00000377022.3 ENST00000344008.5 |
CASZ1 |
castor zinc finger 1 |
| chr9_+_71939488 | 0.18 |
ENST00000455972.1 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr3_-_33686925 | 0.18 |
ENST00000485378.2 ENST00000313350.6 ENST00000487200.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr18_-_53257027 | 0.18 |
ENST00000568740.1 ENST00000564403.2 ENST00000537578.1 |
TCF4 |
transcription factor 4 |
| chr6_-_143771799 | 0.18 |
ENST00000237283.8 |
ADAT2 |
adenosine deaminase, tRNA-specific 2 |
| chrX_+_138612889 | 0.18 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr6_-_28554977 | 0.18 |
ENST00000452236.2 |
SCAND3 |
SCAN domain containing 3 |
| chr6_+_25279651 | 0.17 |
ENST00000329474.6 |
LRRC16A |
leucine rich repeat containing 16A |
| chr8_-_124054484 | 0.17 |
ENST00000419562.2 |
DERL1 |
derlin 1 |
| chr2_+_148602058 | 0.17 |
ENST00000241416.7 ENST00000535787.1 ENST00000404590.1 |
ACVR2A |
activin A receptor, type IIA |
| chr8_-_124054362 | 0.17 |
ENST00000405944.3 |
DERL1 |
derlin 1 |
| chr19_+_41281416 | 0.16 |
ENST00000597140.1 |
MIA |
melanoma inhibitory activity |
| chr8_-_124054587 | 0.16 |
ENST00000259512.4 |
DERL1 |
derlin 1 |
| chr2_-_238499725 | 0.16 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr5_+_35856951 | 0.16 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr17_+_3539998 | 0.16 |
ENST00000452111.1 ENST00000574776.1 ENST00000441220.2 ENST00000414524.2 |
CTNS |
cystinosin, lysosomal cystine transporter |
| chr4_-_80994619 | 0.15 |
ENST00000404191.1 |
ANTXR2 |
anthrax toxin receptor 2 |
| chr8_+_75736761 | 0.15 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr19_-_13227463 | 0.15 |
ENST00000437766.1 ENST00000221504.8 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr20_+_55204351 | 0.15 |
ENST00000201031.2 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr5_+_140174429 | 0.15 |
ENST00000520672.2 ENST00000378132.1 ENST00000526136.1 |
PCDHA2 |
protocadherin alpha 2 |
| chr14_-_93214915 | 0.15 |
ENST00000553918.1 ENST00000555699.1 ENST00000553802.1 ENST00000554397.1 ENST00000554919.1 ENST00000554080.1 ENST00000553371.1 |
LGMN |
legumain |
| chr19_+_41281282 | 0.15 |
ENST00000263369.3 |
MIA |
melanoma inhibitory activity |
| chr2_-_69180083 | 0.15 |
ENST00000328895.4 |
GKN2 |
gastrokine 2 |
| chr1_+_81771806 | 0.14 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr14_-_93214988 | 0.14 |
ENST00000557434.1 ENST00000393218.2 ENST00000334869.4 |
LGMN |
legumain |
| chr6_+_45296391 | 0.14 |
ENST00000371436.6 ENST00000576263.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr12_+_41831485 | 0.14 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr6_+_29364416 | 0.14 |
ENST00000383555.2 |
OR12D2 |
olfactory receptor, family 12, subfamily D, member 2 (gene/pseudogene) |
| chr19_-_7797045 | 0.14 |
ENST00000328853.5 |
CLEC4G |
C-type lectin domain family 4, member G |
| chr15_+_92396920 | 0.13 |
ENST00000318445.6 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
| chr4_-_90756769 | 0.13 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr6_-_10412600 | 0.13 |
ENST00000379608.3 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr12_+_113354341 | 0.13 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr19_-_13227534 | 0.13 |
ENST00000588229.1 ENST00000357720.4 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr19_+_41281060 | 0.13 |
ENST00000594436.1 ENST00000597784.1 |
MIA |
melanoma inhibitory activity |
| chr2_+_153574428 | 0.13 |
ENST00000326446.5 |
ARL6IP6 |
ADP-ribosylation-like factor 6 interacting protein 6 |
| chr6_+_31795506 | 0.13 |
ENST00000375650.3 |
HSPA1B |
heat shock 70kDa protein 1B |
| chr13_-_46756351 | 0.13 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr8_-_66701319 | 0.12 |
ENST00000379419.4 |
PDE7A |
phosphodiesterase 7A |
| chr17_-_41836153 | 0.12 |
ENST00000301691.2 |
SOST |
sclerostin |
| chr8_+_42010464 | 0.12 |
ENST00000518421.1 ENST00000174653.3 ENST00000396926.3 ENST00000521280.1 ENST00000522288.1 |
AP3M2 |
adaptor-related protein complex 3, mu 2 subunit |
| chr4_-_103266626 | 0.12 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr17_-_39280419 | 0.12 |
ENST00000394014.1 |
KRTAP4-12 |
keratin associated protein 4-12 |
| chr2_-_9143786 | 0.12 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr5_+_147582348 | 0.12 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr11_-_117748138 | 0.12 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr2_-_160472952 | 0.12 |
ENST00000541068.2 ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr13_+_47127322 | 0.12 |
ENST00000389798.3 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr17_-_39580775 | 0.12 |
ENST00000225550.3 |
KRT37 |
keratin 37 |
| chr9_-_34376851 | 0.12 |
ENST00000297625.7 |
KIAA1161 |
KIAA1161 |
| chr7_-_71912046 | 0.12 |
ENST00000395276.2 ENST00000431984.1 |
CALN1 |
calneuron 1 |
| chr7_+_130020932 | 0.12 |
ENST00000484324.1 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
| chr12_-_95510743 | 0.12 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr7_-_121944491 | 0.12 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr8_+_118532937 | 0.12 |
ENST00000297347.3 |
MED30 |
mediator complex subunit 30 |
| chr2_+_105050794 | 0.12 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr2_+_234104079 | 0.12 |
ENST00000417661.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr20_-_23066953 | 0.11 |
ENST00000246006.4 |
CD93 |
CD93 molecule |
| chr11_+_59522532 | 0.11 |
ENST00000337979.4 ENST00000535361.1 |
STX3 |
syntaxin 3 |
| chr2_-_160473114 | 0.11 |
ENST00000392783.2 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr14_+_22771851 | 0.11 |
ENST00000390466.1 |
TRAV39 |
T cell receptor alpha variable 39 |
| chr11_+_2323236 | 0.11 |
ENST00000182290.4 |
TSPAN32 |
tetraspanin 32 |
| chr7_+_23636992 | 0.11 |
ENST00000307471.3 ENST00000409765.1 |
CCDC126 |
coiled-coil domain containing 126 |
| chr10_-_13523073 | 0.11 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr2_+_11696464 | 0.11 |
ENST00000234142.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr11_-_62752162 | 0.11 |
ENST00000458333.2 ENST00000421062.2 |
SLC22A6 |
solute carrier family 22 (organic anion transporter), member 6 |
| chr3_-_66024213 | 0.11 |
ENST00000483466.1 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr5_+_176784837 | 0.11 |
ENST00000408923.3 |
RGS14 |
regulator of G-protein signaling 14 |
| chr2_+_162272605 | 0.11 |
ENST00000389554.3 |
TBR1 |
T-box, brain, 1 |
| chr6_+_42584847 | 0.11 |
ENST00000372883.3 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr20_+_43935474 | 0.11 |
ENST00000372743.1 ENST00000372741.3 ENST00000343694.3 |
RBPJL |
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr1_-_155947951 | 0.11 |
ENST00000313695.7 ENST00000497907.1 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr3_-_195619579 | 0.11 |
ENST00000428187.1 |
TNK2 |
tyrosine kinase, non-receptor, 2 |
| chr3_-_119813264 | 0.11 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr4_+_56815102 | 0.11 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
| chr14_-_95624227 | 0.11 |
ENST00000526495.1 |
DICER1 |
dicer 1, ribonuclease type III |
| chr18_-_53177984 | 0.10 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chrX_-_138914394 | 0.10 |
ENST00000327569.3 ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C |
ATPase, class VI, type 11C |
| chr13_-_103719196 | 0.10 |
ENST00000245312.3 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr8_+_133931648 | 0.10 |
ENST00000519178.1 ENST00000542445.1 |
TG |
thyroglobulin |
| chr11_+_93861993 | 0.10 |
ENST00000227638.3 ENST00000436171.2 |
PANX1 |
pannexin 1 |
| chr11_-_72433346 | 0.09 |
ENST00000334211.8 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr15_+_42131011 | 0.09 |
ENST00000458483.1 |
PLA2G4B |
phospholipase A2, group IVB (cytosolic) |
| chr8_+_95835438 | 0.09 |
ENST00000521860.1 ENST00000519457.1 ENST00000519053.1 ENST00000523731.1 ENST00000447247.1 |
INTS8 |
integrator complex subunit 8 |
| chr17_+_27573875 | 0.09 |
ENST00000225387.3 |
CRYBA1 |
crystallin, beta A1 |
| chr12_+_81101277 | 0.09 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr9_-_112260531 | 0.09 |
ENST00000374541.2 ENST00000262539.3 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
| chr8_+_144451039 | 0.09 |
ENST00000289013.6 |
RHPN1 |
rhophilin, Rho GTPase binding protein 1 |
| chr17_+_39261584 | 0.09 |
ENST00000391415.1 |
KRTAP4-9 |
keratin associated protein 4-9 |
| chr18_-_59854203 | 0.09 |
ENST00000589339.1 ENST00000357637.5 ENST00000585458.1 ENST00000400334.3 ENST00000587134.1 ENST00000585923.1 ENST00000590765.1 ENST00000589720.1 ENST00000588571.1 ENST00000585344.1 |
PIGN |
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr17_+_68071389 | 0.09 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_-_135271260 | 0.09 |
ENST00000265605.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr6_+_21593972 | 0.09 |
ENST00000244745.1 ENST00000543472.1 |
SOX4 |
SRY (sex determining region Y)-box 4 |
| chr18_-_19283649 | 0.09 |
ENST00000584464.1 ENST00000578270.1 |
ABHD3 |
abhydrolase domain containing 3 |
| chr6_-_135271219 | 0.09 |
ENST00000367847.2 ENST00000367845.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr17_+_47448102 | 0.09 |
ENST00000576461.1 |
RP11-81K2.1 |
Uncharacterized protein |
| chr19_+_55477711 | 0.09 |
ENST00000448584.2 ENST00000537859.1 ENST00000585500.1 ENST00000427260.2 ENST00000538819.1 ENST00000263437.6 |
NLRP2 |
NLR family, pyrin domain containing 2 |
| chr17_+_11924129 | 0.09 |
ENST00000353533.5 ENST00000415385.3 |
MAP2K4 |
mitogen-activated protein kinase kinase 4 |
| chr1_-_153348067 | 0.09 |
ENST00000368737.3 |
S100A12 |
S100 calcium binding protein A12 |
| chr12_+_113659234 | 0.09 |
ENST00000551096.1 ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1 |
two pore segment channel 1 |
| chr2_+_27799389 | 0.09 |
ENST00000408964.2 |
C2orf16 |
chromosome 2 open reading frame 16 |
| chr11_+_85956182 | 0.09 |
ENST00000327320.4 ENST00000351625.6 ENST00000534595.1 |
EED |
embryonic ectoderm development |
| chr17_+_3539744 | 0.08 |
ENST00000046640.3 ENST00000381870.3 |
CTNS |
cystinosin, lysosomal cystine transporter |
| chr2_-_231989808 | 0.08 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr1_+_46713404 | 0.08 |
ENST00000371975.4 ENST00000469835.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chr1_+_46713357 | 0.08 |
ENST00000442598.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chr1_+_173793641 | 0.08 |
ENST00000361951.4 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chrX_+_84499081 | 0.08 |
ENST00000276123.3 |
ZNF711 |
zinc finger protein 711 |
| chr1_+_173793777 | 0.08 |
ENST00000239457.5 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr5_+_178368186 | 0.08 |
ENST00000320129.3 ENST00000519564.1 |
ZNF454 |
zinc finger protein 454 |
| chr11_-_94964354 | 0.08 |
ENST00000536441.1 |
SESN3 |
sestrin 3 |
| chr7_+_23637118 | 0.08 |
ENST00000448353.1 |
CCDC126 |
coiled-coil domain containing 126 |
| chr12_+_50017327 | 0.08 |
ENST00000261897.1 |
PRPF40B |
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr4_-_96470108 | 0.08 |
ENST00000513796.1 |
UNC5C |
unc-5 homolog C (C. elegans) |
| chr21_-_36421535 | 0.08 |
ENST00000416754.1 ENST00000437180.1 ENST00000455571.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr6_+_30294612 | 0.08 |
ENST00000440271.1 ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39 |
tripartite motif containing 39 |
| chr10_+_118083919 | 0.08 |
ENST00000333254.3 |
CCDC172 |
coiled-coil domain containing 172 |
| chr19_+_6887571 | 0.08 |
ENST00000250572.8 ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1 |
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr3_-_196910721 | 0.08 |
ENST00000443183.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr6_-_146285455 | 0.08 |
ENST00000367505.2 |
SHPRH |
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr5_-_141338627 | 0.08 |
ENST00000231484.3 |
PCDH12 |
protocadherin 12 |
| chr17_+_68071458 | 0.08 |
ENST00000589377.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr18_-_35145728 | 0.08 |
ENST00000361795.5 ENST00000603232.1 |
CELF4 |
CUGBP, Elav-like family member 4 |
| chr12_-_113658892 | 0.08 |
ENST00000299732.2 ENST00000416617.2 |
IQCD |
IQ motif containing D |
| chr6_+_30457244 | 0.07 |
ENST00000376630.4 |
HLA-E |
major histocompatibility complex, class I, E |
| chr6_+_160327974 | 0.07 |
ENST00000252660.4 |
MAS1 |
MAS1 oncogene |
| chr16_-_70323422 | 0.07 |
ENST00000261772.8 |
AARS |
alanyl-tRNA synthetase |
| chr10_+_6392278 | 0.07 |
ENST00000391437.1 |
DKFZP667F0711 |
DKFZP667F0711 |
| chr1_-_115238207 | 0.07 |
ENST00000520113.2 ENST00000369538.3 ENST00000353928.6 |
AMPD1 |
adenosine monophosphate deaminase 1 |
| chr2_-_69180012 | 0.07 |
ENST00000481498.1 |
GKN2 |
gastrokine 2 |
| chr3_+_1134260 | 0.07 |
ENST00000446702.2 ENST00000539053.1 ENST00000350110.2 |
CNTN6 |
contactin 6 |
| chr5_-_88179302 | 0.07 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr1_+_209859510 | 0.07 |
ENST00000367028.2 ENST00000261465.1 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr3_-_158450475 | 0.07 |
ENST00000237696.5 |
RARRES1 |
retinoic acid receptor responder (tazarotene induced) 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.2 | 0.9 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.3 | GO:0098905 | pulmonary valve formation(GO:0003193) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.5 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.5 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 2.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.2 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.1 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.3 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:1904339 | superior temporal gyrus development(GO:0071109) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:0070997 | neuron death(GO:0070997) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 1.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0032759 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) protection from natural killer cell mediated cytotoxicity(GO:0042270) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:0035910 | N-terminal peptidyl-lysine acetylation(GO:0018076) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 1.1 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.3 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:1903781 | endoplasmic reticulum membrane organization(GO:0090158) positive regulation of cardiac conduction(GO:1903781) |
| 0.0 | 0.2 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.5 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 1.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |